One paper been accepted by IEEE ICME 2025
Title: Hypergraph Self-Supervised Learning for Survival Prediction on Whole Slide Images
Existing methods for whole-slide histopathological image analysis commonly depend on patch-level sampling and deep feature extraction based on ImageNet pre-trained models. However, this strategy frequently fails to fully capture the diversity and complex associations within the tissue microenvironment.
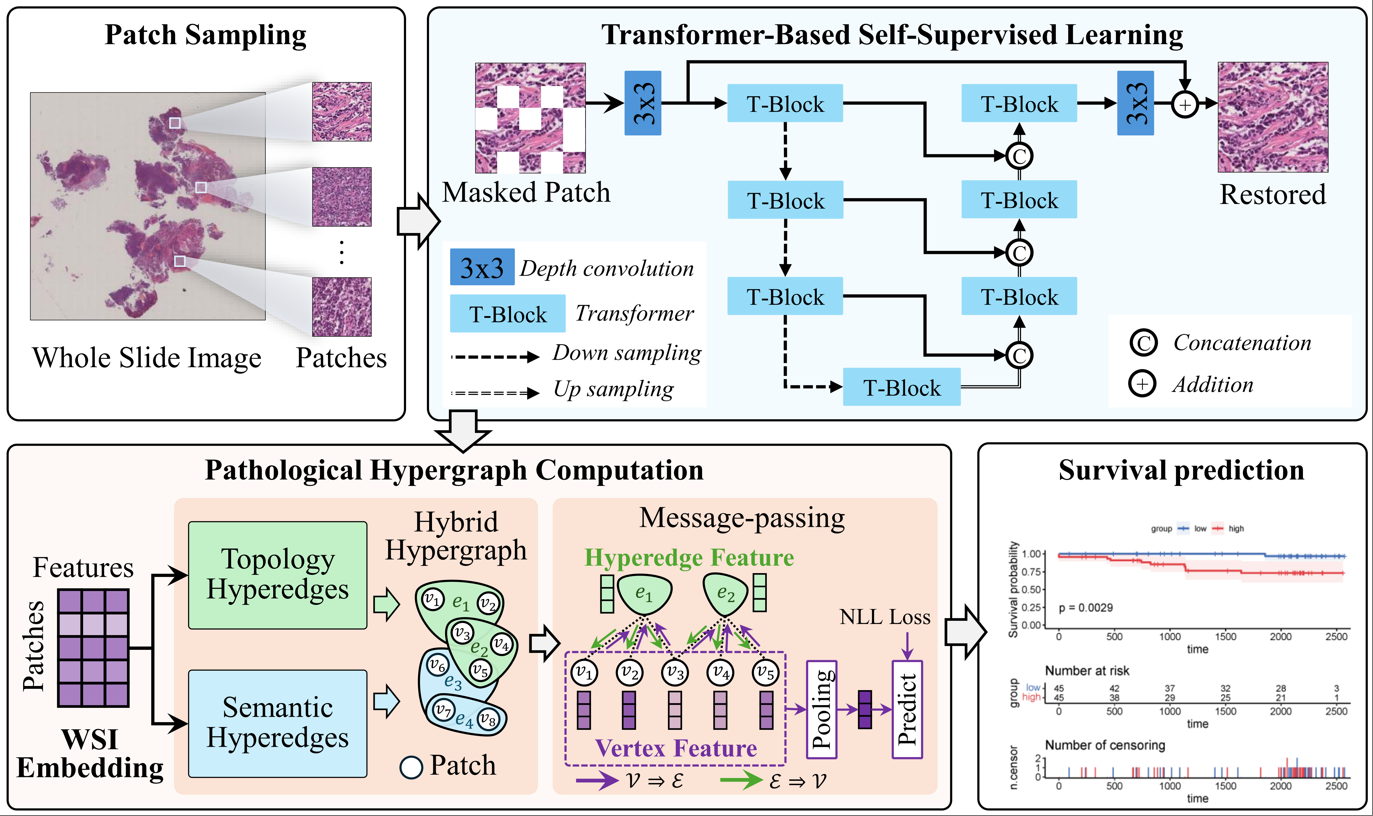
To address this problem, we propose a HyperGraph Self-supervised Learning (HGSL) method to utilize both local and global information in whole-slide images. Specifically, HGSL consists of two key modules: a Transformer-based self-supervised pretraining (TranSSL) module and a pathology hypergraph neural network (PathHGNN) module. TranSSL uses a mask reconstruction task to mine latent structures and patterns from unlabeled data, thus yielding pathology semantic deep representations. PathHGNN builds a hypergraph structure based on these representations to describe high-order relationships among patches, enabling multi-level feature aggregation and propagation to integrate local microenvironmental information with the overall topological correlations.
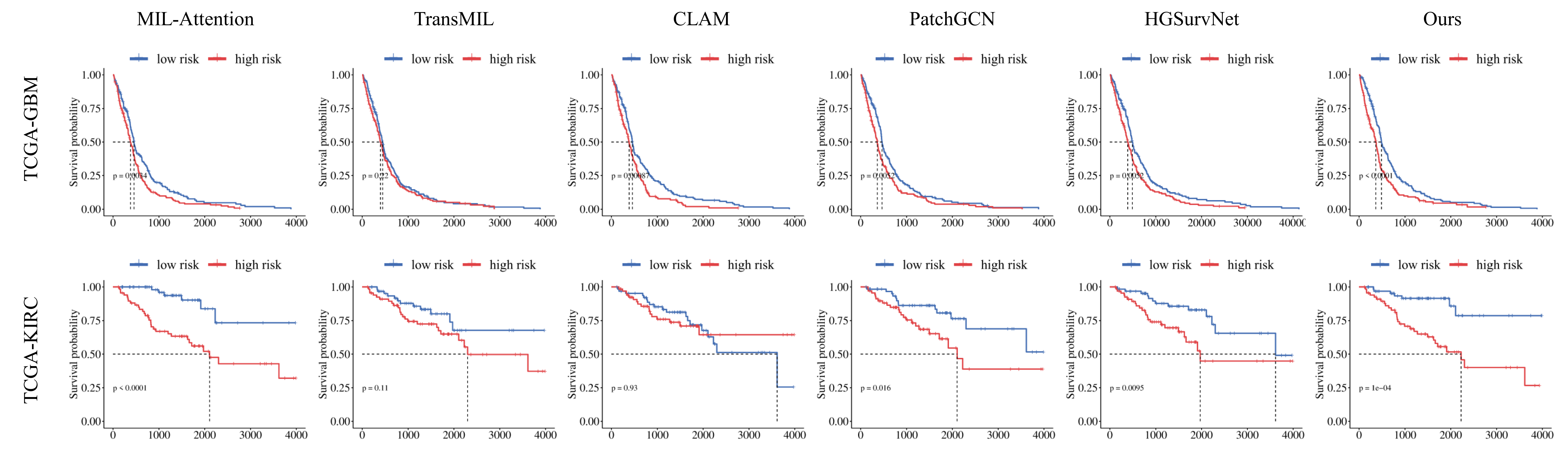
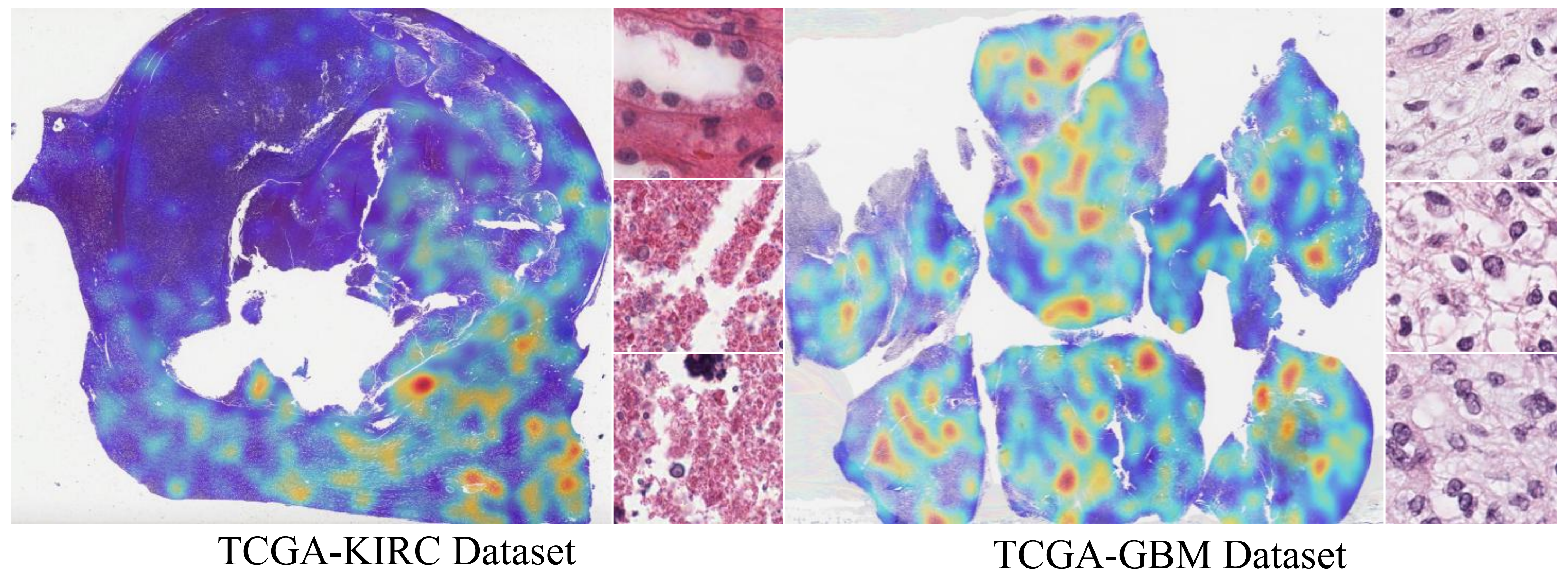
The proposed HGSL was validated on the TCGA-GBM and TCGA-KIRC datasets, and the experimental results show that HGSL significantly outperforms all existing comparative methods in survival prediction tasks. Furthermore, the KM curve demonstrates that the proposed HGSL significantly distinguishes between high- and low-risk groups, indicating its potential for future clinical applications.