One paper been accepted by IEEE TPAMI 🎉
Title: Inter-Intra Hypergraph Computation for Survival Prediction on Whole Slide Images
Lab: https://www.gaoyue.org/
Code: https://github.com/airsimonhan/IIHGC
Survival prediction on histopathology whole slide images (WSIs) involves the analysis of multi-level complex correlations, such as inter-correlations among patients and intra-correlations within gigapixel histopathology images. However, the current graph-based methods for WSI analysis mainly focus on the exploration of pairwise correlations, resulting in the loss of high-order correlations. Hypergraph-based methods can handle such high-order correlations, while existing hypergraph-based methods fail to integrate multi-level high-order correlations into a unified framework, which limits the representation capability of WSIs.
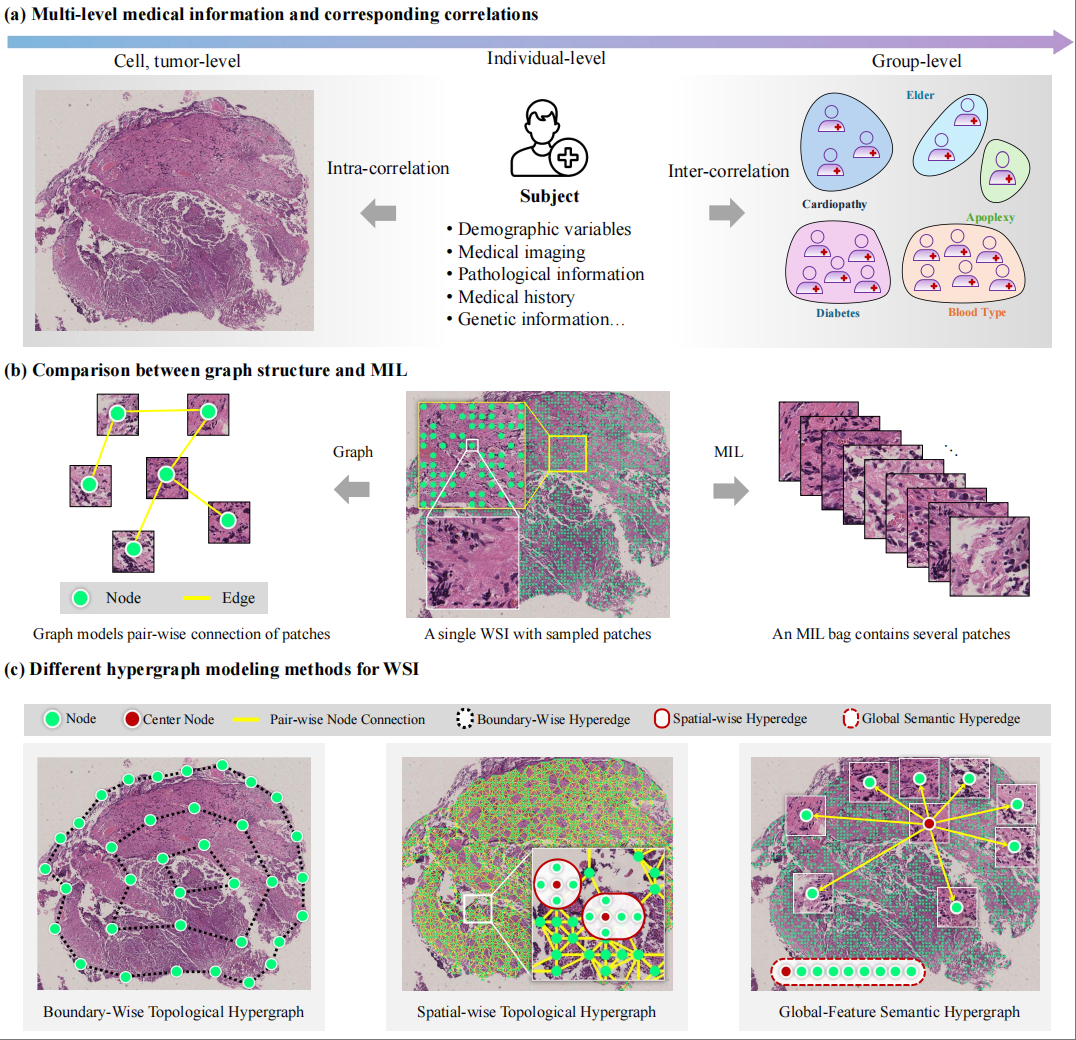
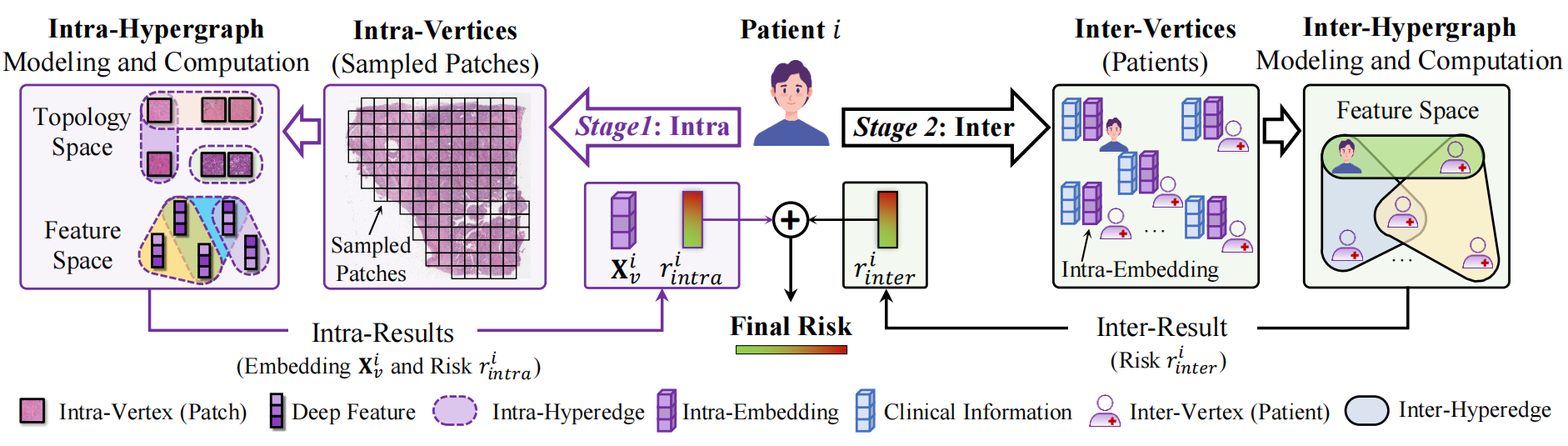
In this work, we propose an inter-intra hypergraph computation ($I^2$HGC) framework to address this issue. The $I^2$HGC framework implements multi-level hypergraph computation for survival prediction on WSIs, namely intra-hypergraph computation and inter-hypergraph computation. Specifically, the intra-hypergraph computation considers each patch sampled from the histopathology WSI as a vertex of the intra-hypergraph and models the high-order correlations among all patches of an individual WSI in both topology and semantic feature spaces using a hypergraph structure. Then, the intra-hypergraph module generates the intra-embedding and intra-risk for each patient. Subsequently, the inter-hypergraph computation employs these intra-embeddings as features for each patient to form the population-level high-order correlations using data- and knowledge-driven hypergraph modeling strategies. Finally, the intra-risks and the inter-risks are fused for the final survival prediction of each patient.
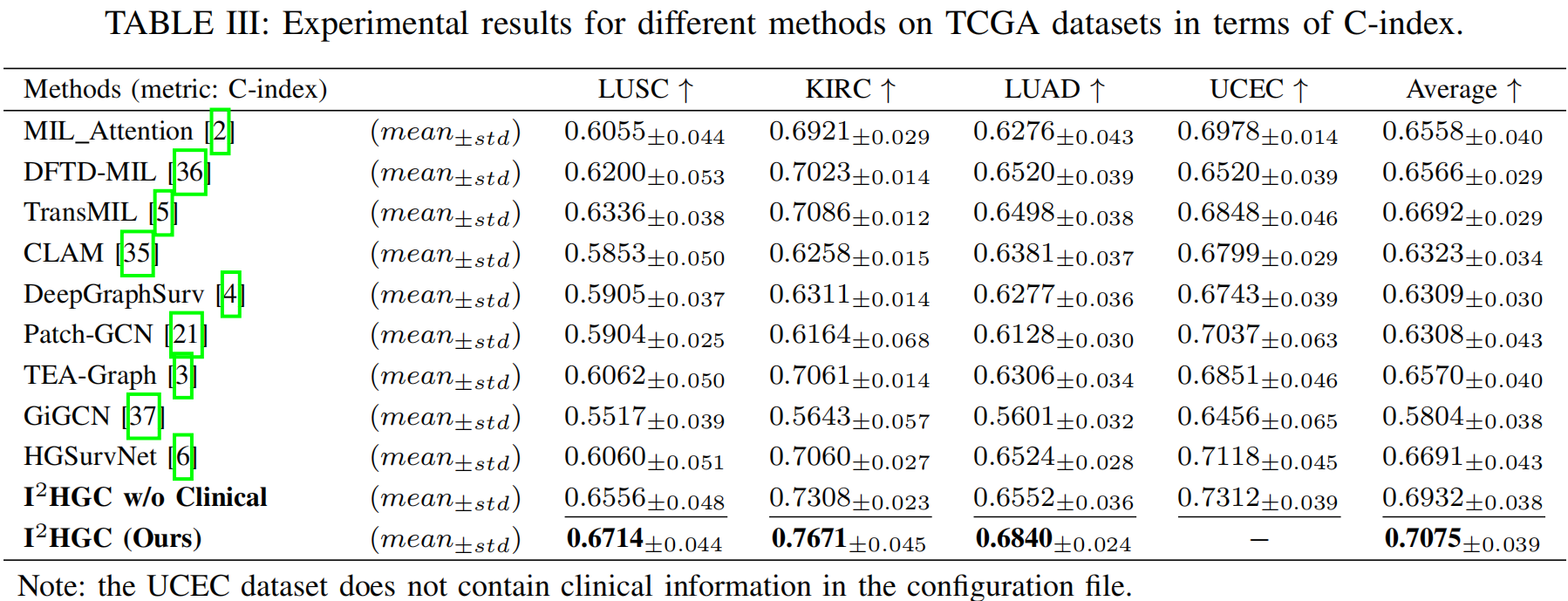
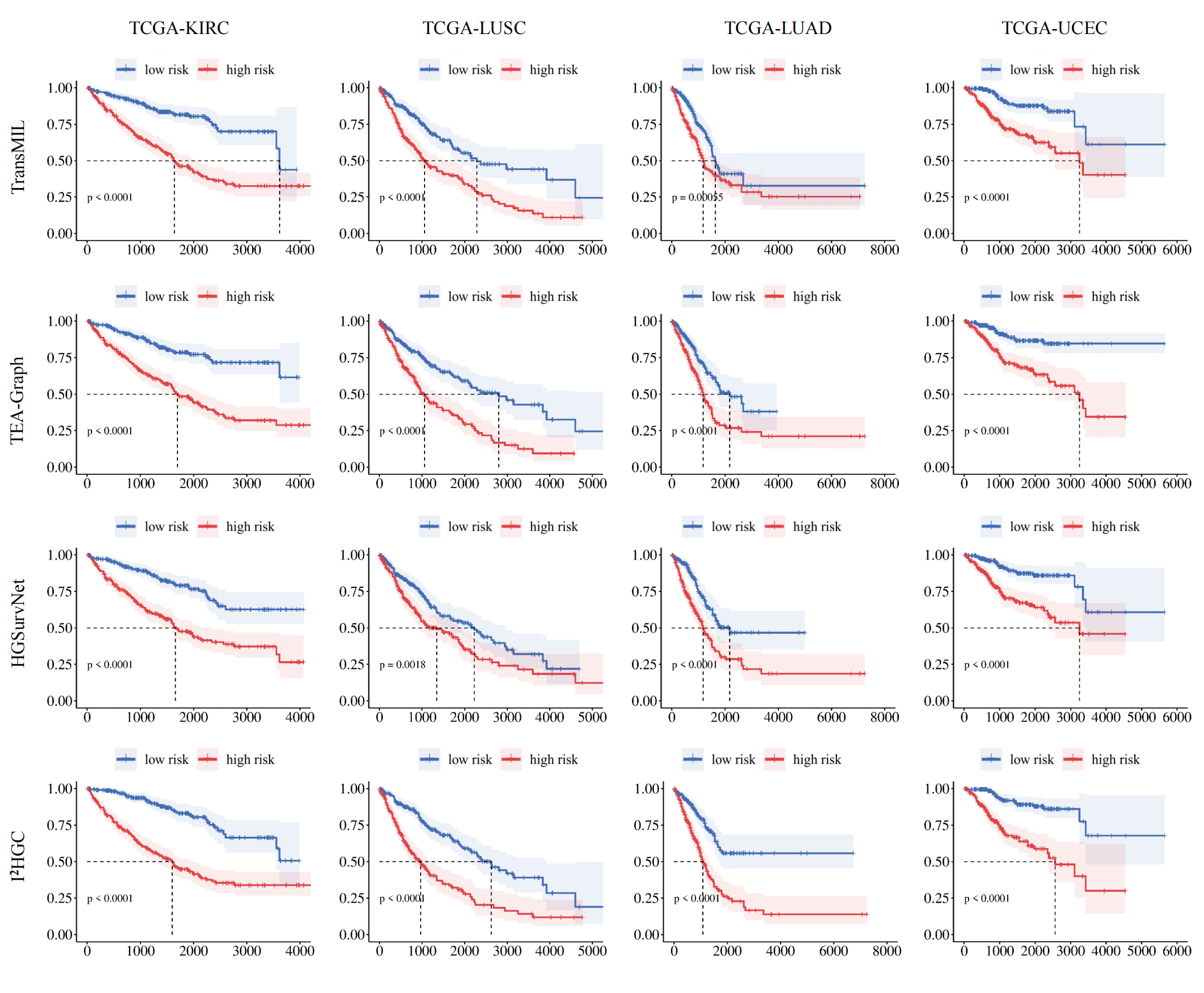
Extensive experimental results on four widely used TCGA carcinoma datasets are presented. We demonstrate that the hypergraph structure captures significantly richer correlations than the graph structure, encompassing all pairwise correlations as well as higher-order interactions through hyperedges. For WSIs with a vast number of pixels and complex correlations, hypergraph-based methods effectively capture topological and semantic information while mitigating the exponential growth of pairwise edges, offering practical advantages for large-scale medical image analysis.
Citation
@article{han_2025_inter,
title = {Inter-intra hypergraph computation for survival prediction on whole slide images},
author = {Han, Xiangmin and Zhou, Huijian and Tian, Zhiqiang and Du, Shaoyi and Gao, Yue},
journal = {IEEE Transactions on Pattern Analysis and Machine Intelligence},
year = {2025},
month = jul,
volume = {47},
number = {7},
pages = {6006--6021},
publisher = {IEEE},
doi = {10.1109/TPAMI.2025.3557391},
}