Four papers been accepted by MICCAI 2025 🎉
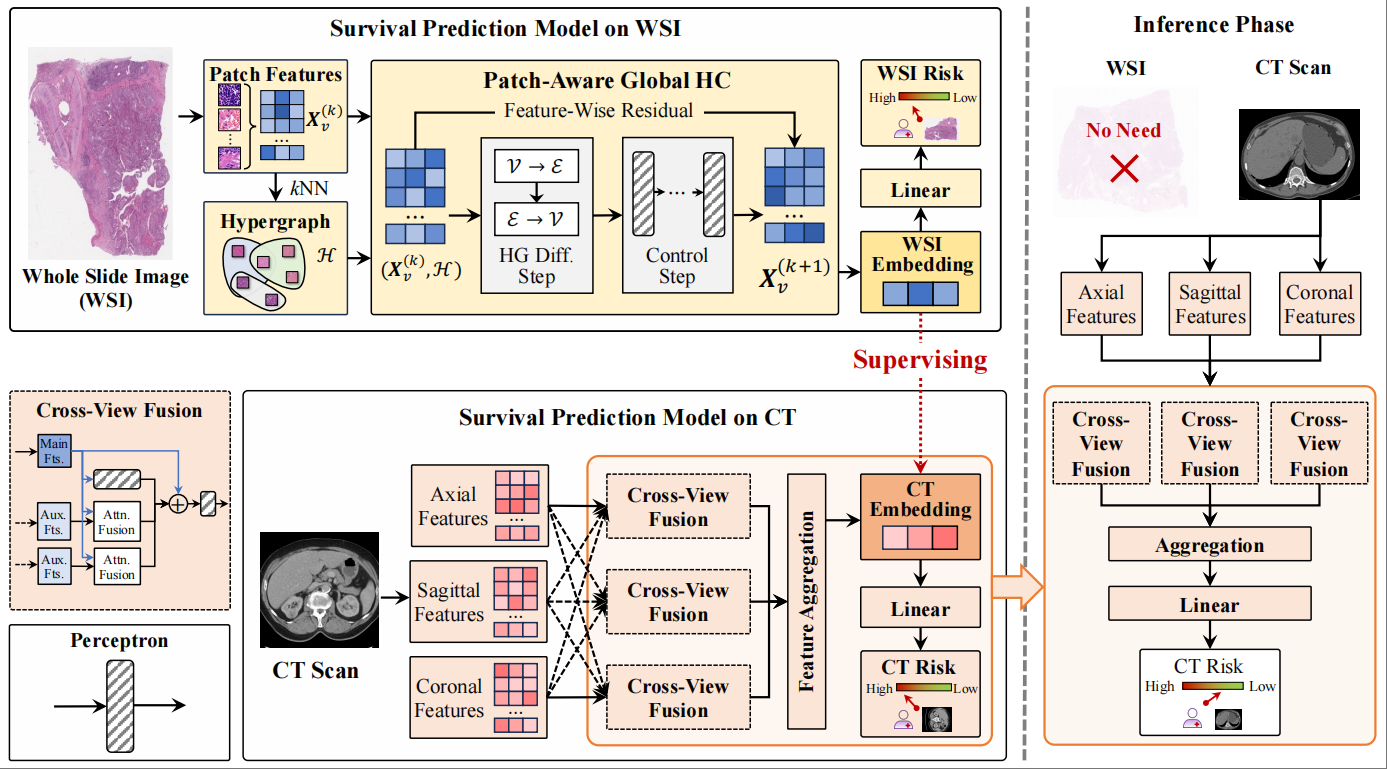
Title: Multimodal Hypergraph Guide Learning for Non-Invasive ccRCC Survival Prediction
Multimodal medical imaging provides critical data for the early diagnosis and clinical management of clear cell renal cell carcinoma (ccRCC). However, early prediction primarily relies on computed tomography (CT), while whole-slide images (WSI) are often unavailable. Consequently, developing a model that can be trained on multimodal data and make predictions using single-modality data is essential. In this paper, we propose a multimodal hypergraph guide learning framework for non-invasive ccRCC survival prediction. First, we propose a patch-aware global hypergraph computation (PAGHC) module, including a hypergraph diffusion step for capturing correlational structure information and a control step to generate stable WSI semantic embeddings. These WSI semantic embeddings are then used to guide a cross-view fusion method, forming the hypergraph WSI-guided cross-view fusion (HWCVF) to generate CT semantic embeddings, improving single-modality performance in inference. We validate our proposed method on three ccRCC datasets, and quantitative results demonstrate a significant improvement in C-index, outperforming state-of-the-art methods.
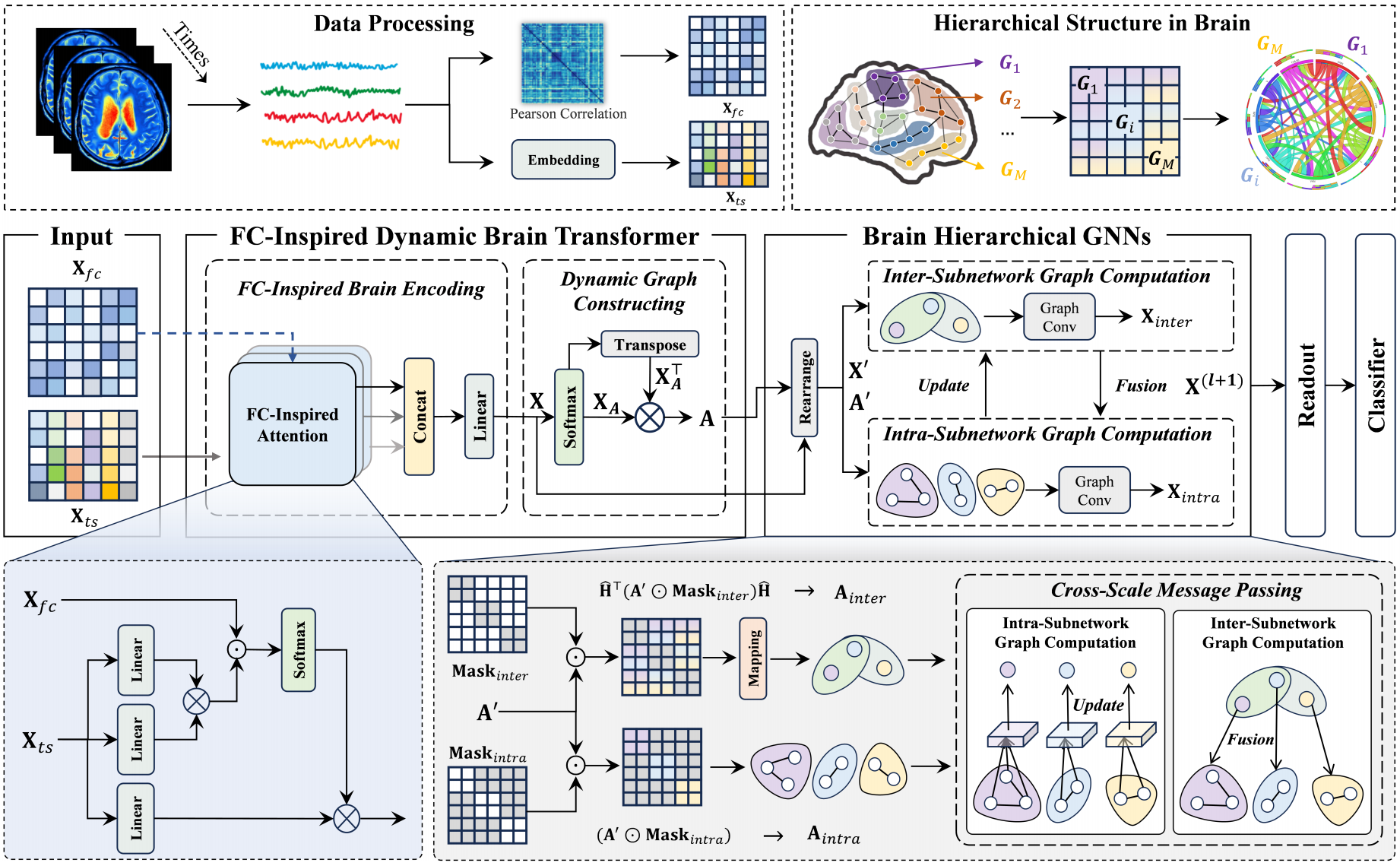
Title: DHGFormer: Dynamic Hierarchical Graph Transformer for Disorder Brain Disease Diagnosis
The functional brain network exhibits a hierarchical characterized organization, balancing localized specialization with global integration through multi-scale hierarchical connectivity. While graph-based methods have advanced brain network analysis, conventional graph neural networks (GNNs) face interpretational limitations when modeling functional connectivity (FC) that encodes excitatory/inhibitory distinctions, often resorting to oversimplified edge weight transformations. Existing methods usually inadequately represent the brain’s hierarchical organization, potentially missing critical information about multi-scale feature interactions. To address these limitations, we propose a novel brain network generation and analysis approach–Dynamic Hierarchical Graph Transformer (DHGFormer). Specifically, our method introduces an FC-driven dynamic attention mechanism that adaptively encodes brain excitatory/inhibitory connectivity patterns into transformer-based representations, enabling dynamic adjustment of the functional brain network. Furthermore, we design hierarchical GNNs that consider prior functional subnetwork knowledge to capture intra-subnetwork homogeneity and inter-subnetwork heterogeneity, thereby enhancing GNN performance in brain disease diagnosis tasks. Extensive experiments on the ABIDE and ADNI datasets demonstrate that DHGFormer consistently outperforms state-of-the-art methods in diagnosing neurological disorders. The code is available at https://anonymous.4open.science/r/MICCAI-DHGFormer.
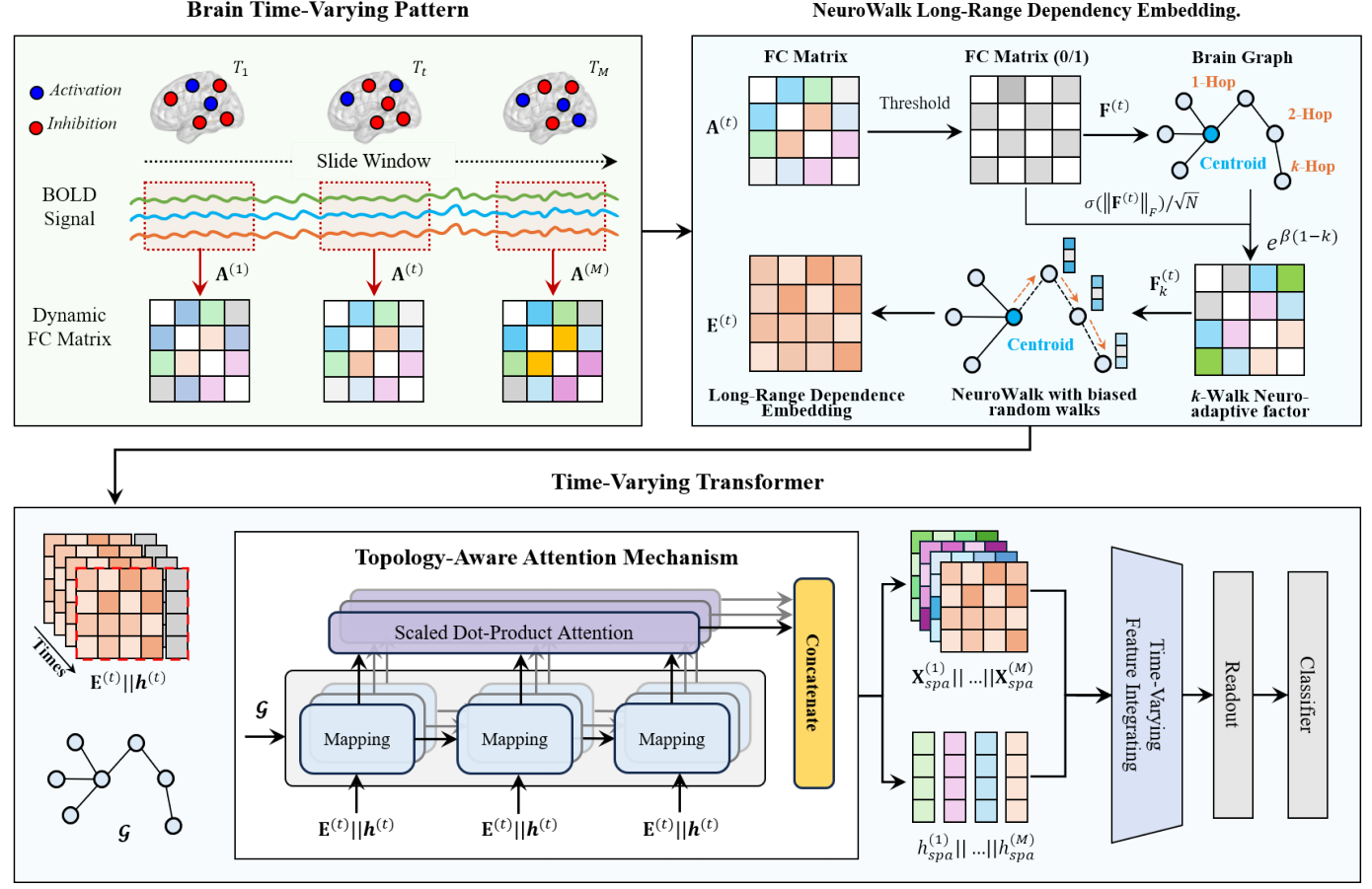
Title: Adaptive Embedding for Long-Range High-Order Dependencies via Time-Varying Transformers on fMRI
Dynamic functional brain network analysis using rs-fMRI has emerged as a powerful approach to understanding brain disorders. However, current methods predominantly focus on pairwise brain region interactions, neglecting critical high-order dependencies and time-varying communication mechanisms. To address these limitations, we propose the Long-Range High-Order Dependency Transformer (LHDFormer), a neurophysiologically-inspired framework that integrates multiscale long-range dependencies with time-varying connectivity patterns. Specifically, we present a biased random walk sampling strategy with NeuroWalk kernel-guided transfer probabilities that dynamically simulate multi-step information loss through a $k$-walk neuroadaptive factor, modeling brain neurobiological principles such as distance-dependent information loss and state-dependent pathway modulation. This enables the adaptive capture of the multi-scale short-range couplings and long-range high-order dependencies corresponding to different steps across evolving connectivity patterns. Complementing this, the time-varying transformer co-embeds local spatial configurations via topology-aware attention and global temporal dynamics through cross-window token guidance, overcoming the single-domain bias of conventional graph/transformer methods. Extensive experiments on ABIDE and ADNI datasets demonstrate that LHDFormer outperforms state-of-the-art methods in brain disease diagnosis. Crucially, the model identifies interpretable high-order connectivity signatures, revealing disrupted long-range integration patterns in patients that align with known neuropathological mechanisms.
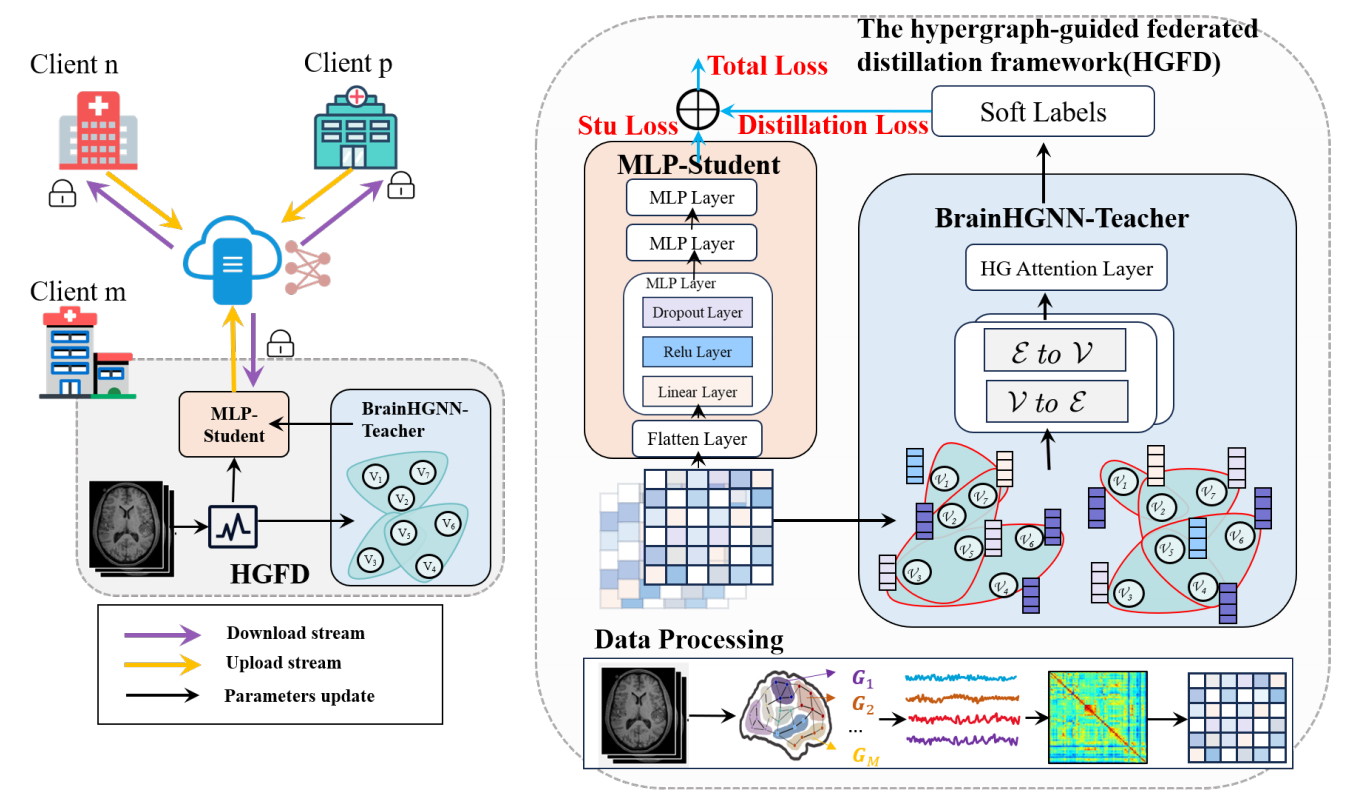
Title: HGFD: Hypergraph Guided Federated Distillation Learning for Efficient and Robust Multi-Center fMRI Data Analysis
Multi-center fMRI data analysis faces significant challenges such as data privacy concerns and data integration issues. Federated learning, as an innovative distributed machine learning approach, enables cross-center collaboration by sharing model parameters instead of raw data. However, existing methods often struggle with improving the robustness and inference efficiency of multi-center fMRI data processing. To address these challenges, we propose a novel hypergraph-guided federated distillation framework(HGFD) for multi-center fMRI data analysis. HGFD utilizes a hypergraph structure to model the spatiotemporal features of brain activity, capturing high-order correlations across brain regions. Additionally, a hypergraph knowledge distillation technique is introduced to distill high-order structural information to shallow neural networks, preserving the ability to infer complex relationships while significantly enhancing inference efficiency. In the federated learning process, participating centers only need to share the parameters of their shallow neural networks to a central server. Through parameter aggregation, each center’s shallow network can learn the high-order structural information of other centers. Experiments on multi-center fMRI datasets demonstrate that the proposed method not only improves the robustness and consistency of fMRI-based prediction tasks but also reduces inference time, achieving efficient and accurate predictions while ensuring data privacy.