MICCAI 2025 Workshop
7th Workshop on GRaphs in biomedicAl Image anaLysis
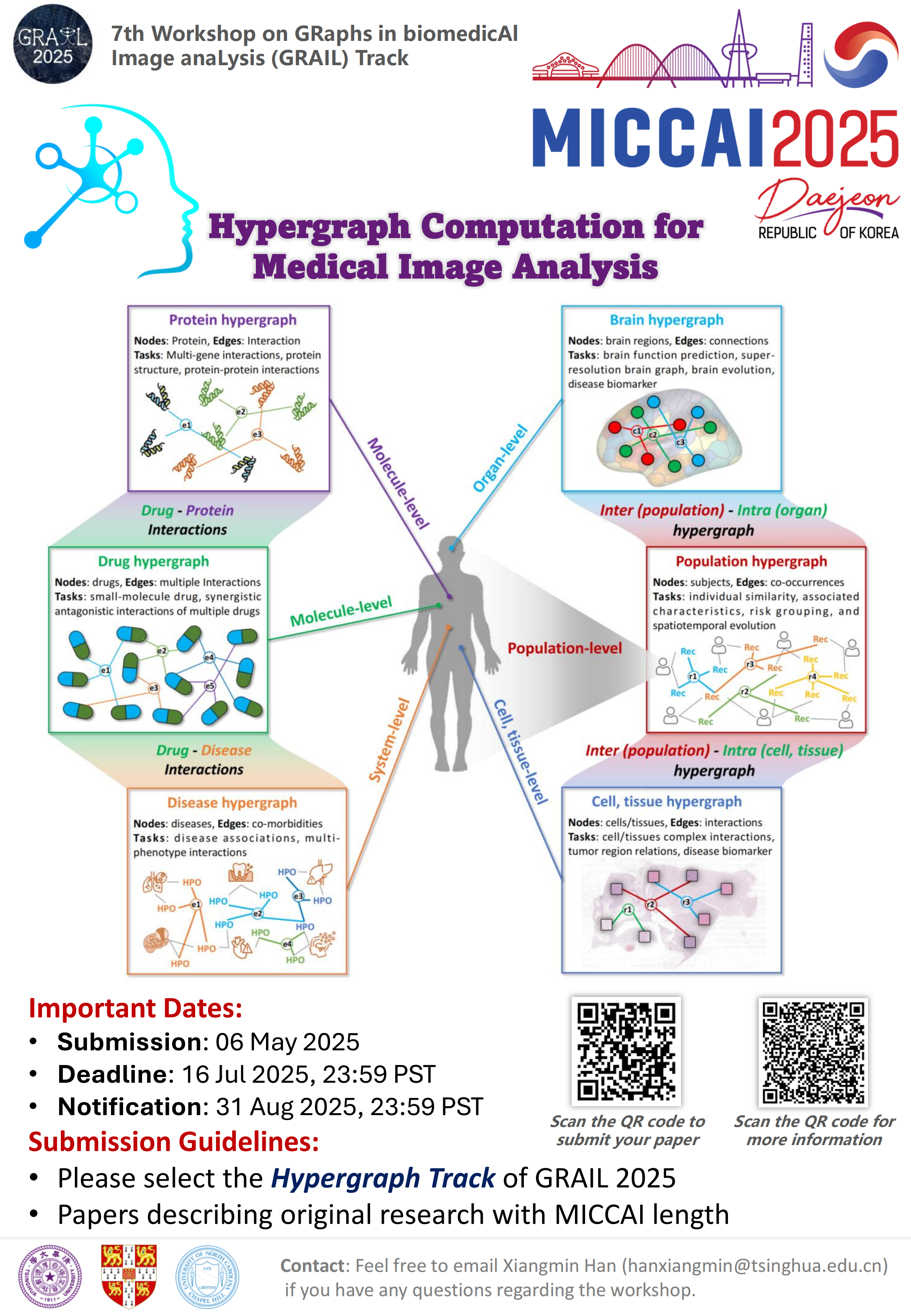
Hypergraph Computation for Medical Image Analysis (HGMIA)
7th Workshop on GRaphs in biomedicAl Image anaLysis (GRAIL) Track
News
- Jun. 25, 2025 We are happy to have Prof. Yue Gao as our keynote speaker. 🥳🥳🥳
- Jun. 19, 2025 Submissions are now open! Submit your paper via the CMT portal.! 🚀🚀🚀
- Jun. 18, 2025 The website is available! 🥳🥳🥳
Schedule
The submission deadlines are in PST timezone.
Introduction
With the continuous advancement of medical imaging technology and the progress in medical data acquisition methods, clinical diagnostics and treatments have generated a vast amount of multi-source heterogeneous medical data. These diverse types of medical data contain critical features related to patients and diseases, driving the development of medical artificial intelligence technologies, particularly in the field of medical image processing. Modeling the complex relationships within and between medical data through algorithmic approaches to achieve more comprehensive, accurate, and diverse feature representations of diseases and patients is a research hotspot in the field of smart healthcare. This is not only a key technology in medical image analysis but also central to subsequent clinical tasks such as auxiliary diagnosis and prognosis evaluation.
Medical data, however, presents unique challenges compared to data from other domains. Due to its high value and the inherent limitations in data collection and usage permissions, data scarcity often poses significant obstacles to medical analysis. Additionally, the intricate and still-understood mechanisms of medical diagnosis and treatment underscore the need to unravel the complex inherent relationships within medical data.
To address these challenges, hypergraph computation has emerged as a promising solution for modeling complex high-order relationships within medical data.
Research Topics:
- High-Order Correlations in Brain Networks: Utilizing hypergraph models to capture complex multi-connection relationships between brain regions, enhancing analysis capabilities of brain functional/structural networks.
- High-Order Correlations in Pathological Images: Using hypergraphs to describe interactions among cells/tissues in whole slide images, improving precision of cancer diagnosis and prognosis assessment.
- Multimodal Medical Image Fusion: Leveraging hypergraph computation to integrate different imaging modalities (MRI, CT, PET) for comprehensive disease representation.
- Cross-Domain Hypergraph Computation: Modeling complex interactions across drugs, genes, diseases and population dimensions for advanced medical analysis.
Keynote
Hypergraph Computation for Medical Image Analysis
Graph learning and graph neural networks have attracted much attention in both research and industrial fields and become very hot topics in these years. It is noted that the world is far more complex than just pairwise connections. Hypergraph, as a generation of graphs, is able to formulate such high-order correlations among the data and has been investigated in last decades. In this part, we first introduce the basic concepts and characteristics of hypergraphs. Next, focusing on hypergraph computation, we introduce hypergraph structural modelling, hypergraph structural evolution, and hypergraph neural network models. Finally, we introduce the application of hypergraph computation for medical data.
Organizers


To contact the organizers, please send email to Xiangmin Han.
Submission
To submit a paper, simply select the Hypergraph Track of GRAIL 2025 in the author console on the GRAIL 2025 CMT portal.
This year we will accept papers in the following format:
-
Complete papers: Papers describing original research with MICCAI length (max 8 pages text + max 2 pages references, supplemental material allowed). Papers are eligible for the “Best Paper Award” and will be published in the GRAIL Open Proceedings and Lecture Notes in Computer Sciences (LNCS) series. Submissions should be anonymous, and formatted following the LNCS Style.
-
All complete papers will be peer-reviewed by 3 members of the program committee, with a double-blinded review process. The selection of the papers will be based on the significance of results, technical merit, relevance, and clarity of presentation.
The Microsoft CMT service was used for managing the peer-reviewing process for the GRAIL 2025 workshop. This service was provided for free by Microsoft and they bore all expenses, including costs for Azure cloud services as well as for software development and support.
More Details