publications
2026
- IEEE TPAMI, IF 18.6
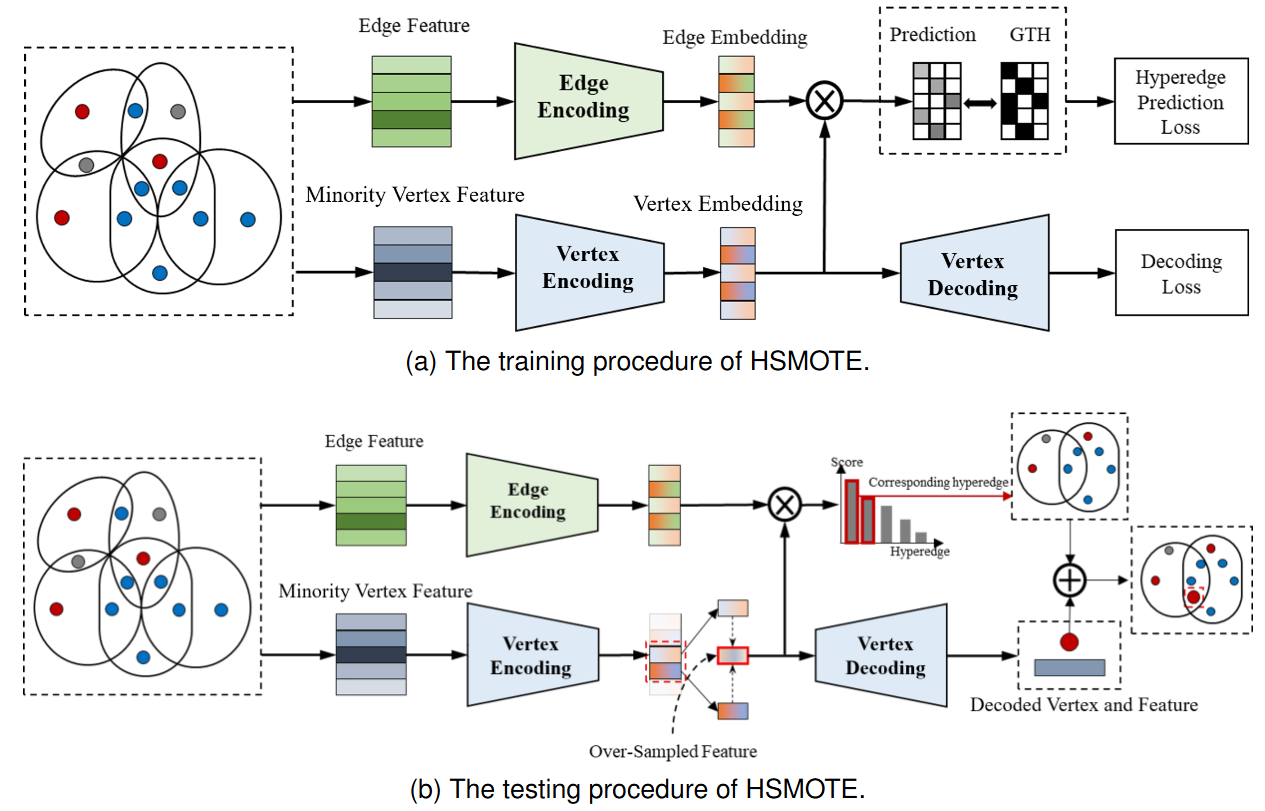 Hypergraph-Based High-Order Correlation Analysis for Large-Scale Long-Tailed Data red ClassificationXiangmin Han, Yubo Zhang, Shihui Ying, and Yue Gao*IEEE Transactions on Pattern Analysis and Machine Intelligence, Jan 2026
Hypergraph-Based High-Order Correlation Analysis for Large-Scale Long-Tailed Data red ClassificationXiangmin Han, Yubo Zhang, Shihui Ying, and Yue Gao*IEEE Transactions on Pattern Analysis and Machine Intelligence, Jan 2026High-order correlations, which capture complex interactions among multiple entities, extend beyond traditional graph representations and support a wider range of applications. However, existing neural network models for high-order correlations encounter scalability issues on large datasets due to the substantial computational complexity involved in processing large-scale structures. In addition, long-tailed distributions, which are common in real-world data, result in underrepresented categories and hinder the model’s ability to learn effective high-order interaction patterns for rare instances. To address these issues, we introduce a novel framework known as HyperGraph-based High-order Correlation analysis (HGHC) for large-scale long-tailed data classification. Firstly, to tackle the long-tailed distribution problem, HGHC generates synthetic vertices and computes their attributed high-order correlations using an oversampling module inspired by SMOTE, termed HSMOTE, to enhance the representation of tail categories. Secondly, for efficient computational scaling, we treat the data as having two modalities: the structural modality capturing high-order relationships and the feature modality representing individual attributes. We perform computations on both CPU and GPU separately and then fuse the results to achieve a lightweight vertex transformation and aggregation scheme for high-order correlation data. Additionally, we contribute the first benchmark for large-scale long-tailed datasets involving high-order correlations, known as Amazon-LT, which includes multiple datasets with varying imbalance ratios. Our experimental results demonstrate that HGHC achieves state-of-the-art performance in handling high-order correlation analysis issues for large-scale, long-tailed data.
@article{han_2025_hghc, title = {Hypergraph-Based High-Order Correlation Analysis for Large-Scale Long-Tailed Data red Classification}, author = {Han, Xiangmin and Zhang, Yubo and Ying, Shihui and Gao, Yue}, journal = {IEEE Transactions on Pattern Analysis and Machine Intelligence}, year = {2026}, month = jan, volume = {48}, number = {1}, pages = {270--282}, publisher = {IEEE}, doi = {10.1109/TPAMI.2025.3603631}, } - IEEE TMM, IF: 9.7
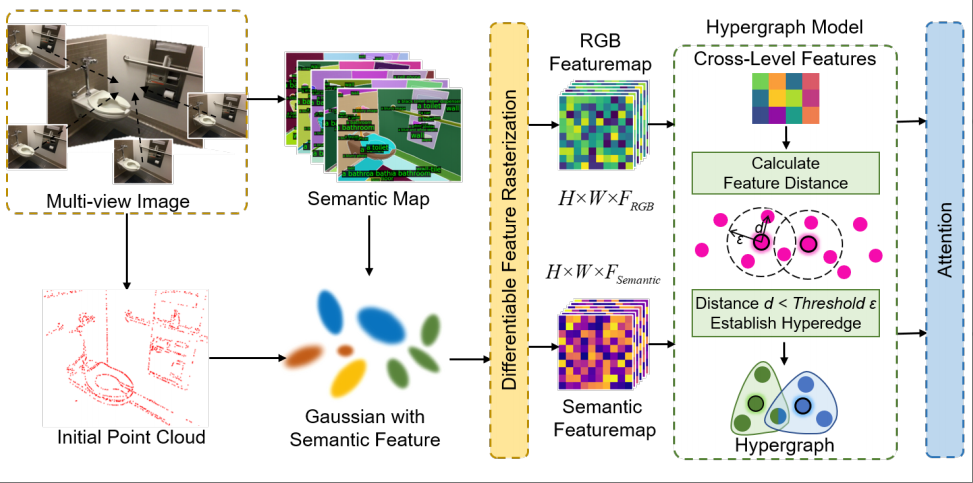 3D Semantic Gaussian via Geometric-Semantic Hypergraph ComputationXinran Wang, Zhiqiang Tian, Dejian Guo, Siqi Li, Shaoyi Du, Xiangmin Han*, and Yue GaoIEEE Transactions on Multimedia, Jun 2026
3D Semantic Gaussian via Geometric-Semantic Hypergraph ComputationXinran Wang, Zhiqiang Tian, Dejian Guo, Siqi Li, Shaoyi Du, Xiangmin Han*, and Yue GaoIEEE Transactions on Multimedia, Jun 2026Semantic labels are inherently tied to geometry and luminance reconstruction, as entities with similar shapes and appearances often share categories. Traditional methods use synthesis-analysis, NeRF, or 3D Gaussian representations to encode semantics and geometry separately. However, 2D methods lack view consistency, NeRF extensions are slow, and faster 3D Gaussian methods risk spatial and channel inconsistencies between semantic and RGB. Moreover, these methods require costly manual dense semantic labels. To alleviate resource demands and achieve effective semantic reconstruction with sparse inputs while enhancing RGB rendering quality, we build upon 3D Gaussian by integrating semantic features from pre-trained models—requiring no additional ground truth input—into Gaussian features, and construct a hypergraph neural network to capture higher-order correlations across RGB and semantic information as well as between different frames. Hypergraphs use hyperedges to link multiple vertices, capturing complex relationships essential for cross-modal tasks. This higher-order structure addresses the limitations of NeRF and Gaussian methods, which lack the capacity for such advanced associations. This framework enables precise novel view synthesis and 2D semantic reconstruction without manual annotations, achieving state-of-the-art results for RGB and semantic tasks on room-scale scenes in the ScanNet and Replica datasets, while supporting real-time rendering speeds of 34 FPS.
@article{han_2025_gshc, title = {3D Semantic Gaussian via Geometric-Semantic Hypergraph Computation}, author = {Wang, Xinran and Tian, Zhiqiang and Guo, Dejian and Li, Siqi and Du, Shaoyi and Han, Xiangmin and Gao, Yue}, journal = {IEEE Transactions on Multimedia}, year = {2026}, month = jun, doi = {10.1109/TMM.2026.3651112}, } - PR, IF: 7.5
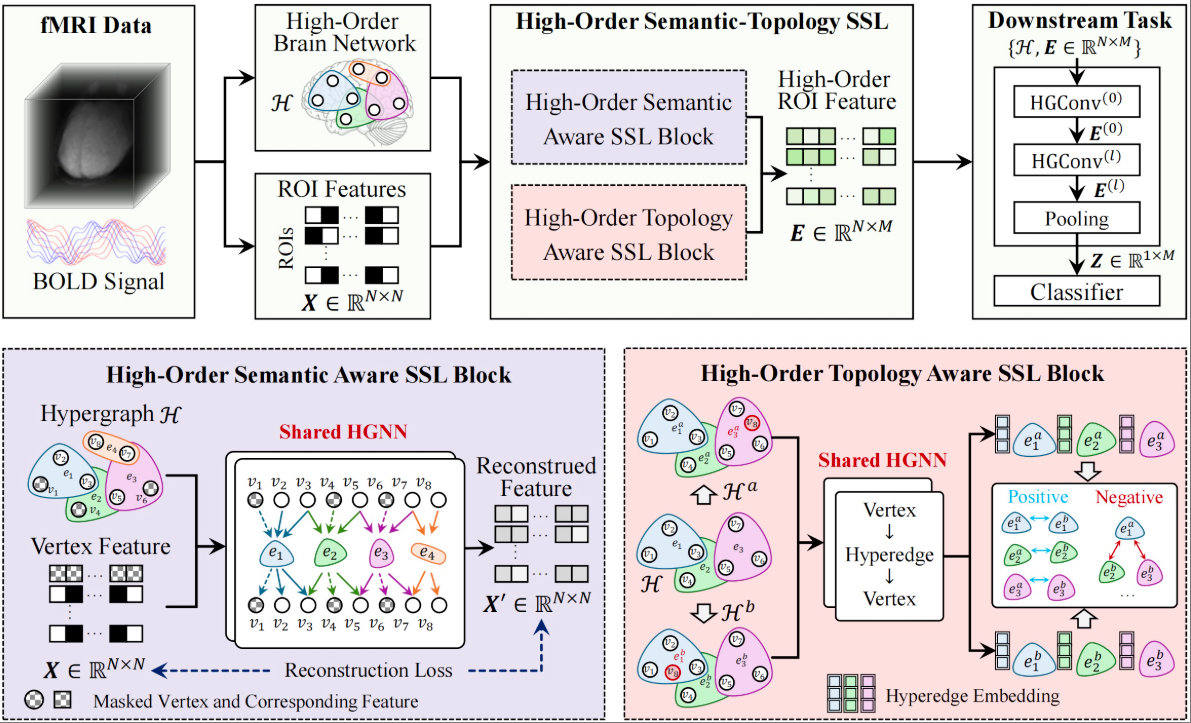 Hypergraph-Based Semantic and Topological Self-Supervised Learning for Brain Disease DiagnosisXiangmin Han, Mengqi Lei, and Junchang Li*Pattern Recognition, Jan 2026
Hypergraph-Based Semantic and Topological Self-Supervised Learning for Brain Disease DiagnosisXiangmin Han, Mengqi Lei, and Junchang Li*Pattern Recognition, Jan 2026Brain networks exhibit complex multi-view interactions, and accurately modeling these interactions is crucial for enhancing the accuracy of brain disease diagnosis. However, existing self-supervised learning methods often overlook the high-order topological and semantic information within brain networks, limiting the performance of pretrained model embeddings. To address this critical issue, we propose a Hypergraph-Based Semantic and Topological Self-Supervised Learning method (HGST) for brain disease diagnosis. This approach leverages hypergraph structures to model high-order functional brain networks and employs hypergraph self-supervised learning to extract latent topological and semantic information from these networks, generating high-order embedding representations that are ultimately applied to downstream brain disease diagnostic tasks. Specifically, HGST consists of two core components: a high-order semantic-aware module based on hypergraph link prediction and a high-order topology-aware module based on hyperedge structure similarity measurement. The former learns high-order embedding of brain regions by restoring masked node features, while the latter captures local topological associations within the brain network by measuring the similarity of hyperedges composed of multiple brain regions. Finally, we utilize the pretrained shared hypergraph neural network as an encoder for downstream tasks, extracting high-order brain network embeddings for each subject to assist in brain disease diagnosis. This method was validated on two public brain disease datasets, ADHD and MDD, demonstrating that HGST significantly outperforms existing methods in diagnostic performance. Additionally, we visualized the high-order brain networks generated by hypergraph self-supervised learning and identified key biomarkers related to the diseases in both ADHD and MDD, further validating the effectiveness and interpretability of our approach.
@article{han_2025_hgst, title = {Hypergraph-Based Semantic and Topological Self-Supervised Learning for Brain Disease Diagnosis}, author = {Han, Xiangmin and Lei, Mengqi and Li, Junchang}, journal = {Pattern Recognition}, year = {2026}, month = jan, volume = {169}, pages = {111921}, doi = {10.1016/j.patcog.2025.111921}, } - IEEE TPAMI, IF 18.6
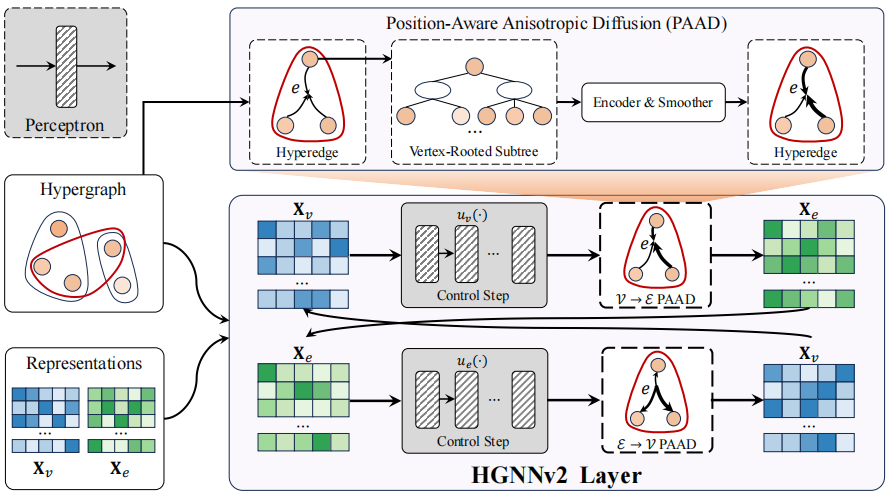 HGNNv2: Stable Hypergraph Neural NetworksYue Gao, Jielong Yan, Yifan Feng, Xiangmin Han, Shihui Ying, Zongze Wu, and Han HuIEEE Transactions on Pattern Analysis and Machine Intelligence, Jan 2026
HGNNv2: Stable Hypergraph Neural NetworksYue Gao, Jielong Yan, Yifan Feng, Xiangmin Han, Shihui Ying, Zongze Wu, and Han HuIEEE Transactions on Pattern Analysis and Machine Intelligence, Jan 2026Hypergraph neural networks (HGNNs) are widely used models for analyzing higher-order relational data. HGNNs suffer from the rapid performance degradation with increasing layers. Hypergraph dynamic system (HDS) is a potential way to deal with this challenge. However, hypergraph dynamic system is confined to a time-continuous isotropic model, lacking positional information in the structural space of the hypergraph. In contrast, anisotropic diffusion can capture structural space differences among vertices, providing a more precise representation of the information propagation process in hypergraph structures than isotropic diffusion. In this paper, we introduce HGNNv2, a stable hypergraph neural network, which is built as a hypergraph dynamic system with partial differential equation (PDE). This model incorporates a position-aware anisotropic diffusion term and an external control term. We further present the vertex-rooted subtree method to determine anisotropic diffusion intensity. HGNNv2 has properties that vertices occupying equivalent positions in the structural space share equivalent structural labels and positional features. Experiments on 6 hypergraph datasets and 3 graph datasets reveal that HGNNv2 outperforms all 12 compared methods. HGNNv2 is capable of achieving stable final representations and task accuracy even under noisy conditions. HGNNv2 achieves stable performance with fewer layers than hypergraph dynamic systems employing isotropic diffusion. We provide feature visualizations to illustrate the evolution of representations.
- AAAI 2026
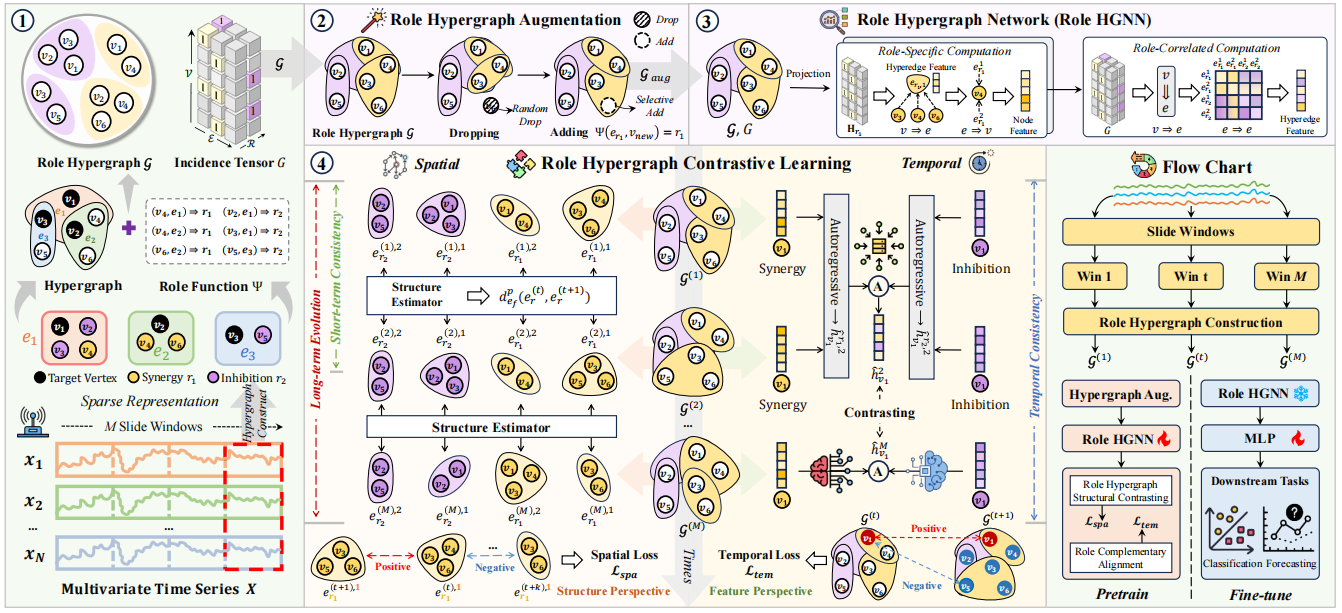 Role Hypergraph Contrastive Learning for Multivariate Time-Series AnalysisRundong Xue, Hao Hu, Zhitao Zeng, Xiangmin Han, Zhiqiang Tian, Yue Gao*, and Shaoyi Du*In AAAI Conference on Artificial Intelligence, Jan 2026
Role Hypergraph Contrastive Learning for Multivariate Time-Series AnalysisRundong Xue, Hao Hu, Zhitao Zeng, Xiangmin Han, Zhiqiang Tian, Yue Gao*, and Shaoyi Du*In AAAI Conference on Artificial Intelligence, Jan 2026Multivariate Time-Series (MTS) analysis is crucial across various domains. Considering the spatial and temporal consistency of MTS, existing methods leverage graph structures with temporal augmentation and contrastive learning to achieve robust learning of spatial dependencies and temporal patterns. Given the inherent high-order correlations in MTS, hypergraphs present a promising approach. However, two key challenges limit their further development: 1) Feature-based perspectives capture limited spatial information, while structural perspectives encode richer spatial consistency and evolution dependency; 2) Various semantic patterns (e.g., synergy, inhibition) entangle in sensor correlations, leading to semantic ambiguity. The underlying reason is that conventional hypergraph structures cannot distinguish specific semantic roles within or across hyperedges. Thus, we propose Role Hypergraph Contrastive Learning for MTS analysis. Specifically, we introduce the concept of role to generalize hypergraphs to Role Hypergraphs, enabling precise modeling of sensor correlations by assigning each vertex-hyperedge pair with a semantic role. Building on this structure, we design a role hypergraph contrastive learning paradigm to comprehensively capture the spatial and temporal dependencies: From a structural perspective, role hypergraph structural contrasting captures spatial short-term consistency and long-term evolution; from a feature perspective, alignment of complementary role information ensures sensor-level temporal consistency. Experiments on classification and forecasting tasks demonstrate the effectiveness and interpretability of our method.
@inproceedings{xue2026role, title = {Role Hypergraph Contrastive Learning for Multivariate Time-Series Analysis}, author = {Xue, Rundong and Hu, Hao and Zeng, Zhitao and Han, Xiangmin and Tian, Zhiqiang and Gao, Yue and Du, Shaoyi}, booktitle = {AAAI Conference on Artificial Intelligence}, year = {2026}, }
2025
- MICCAI 2025
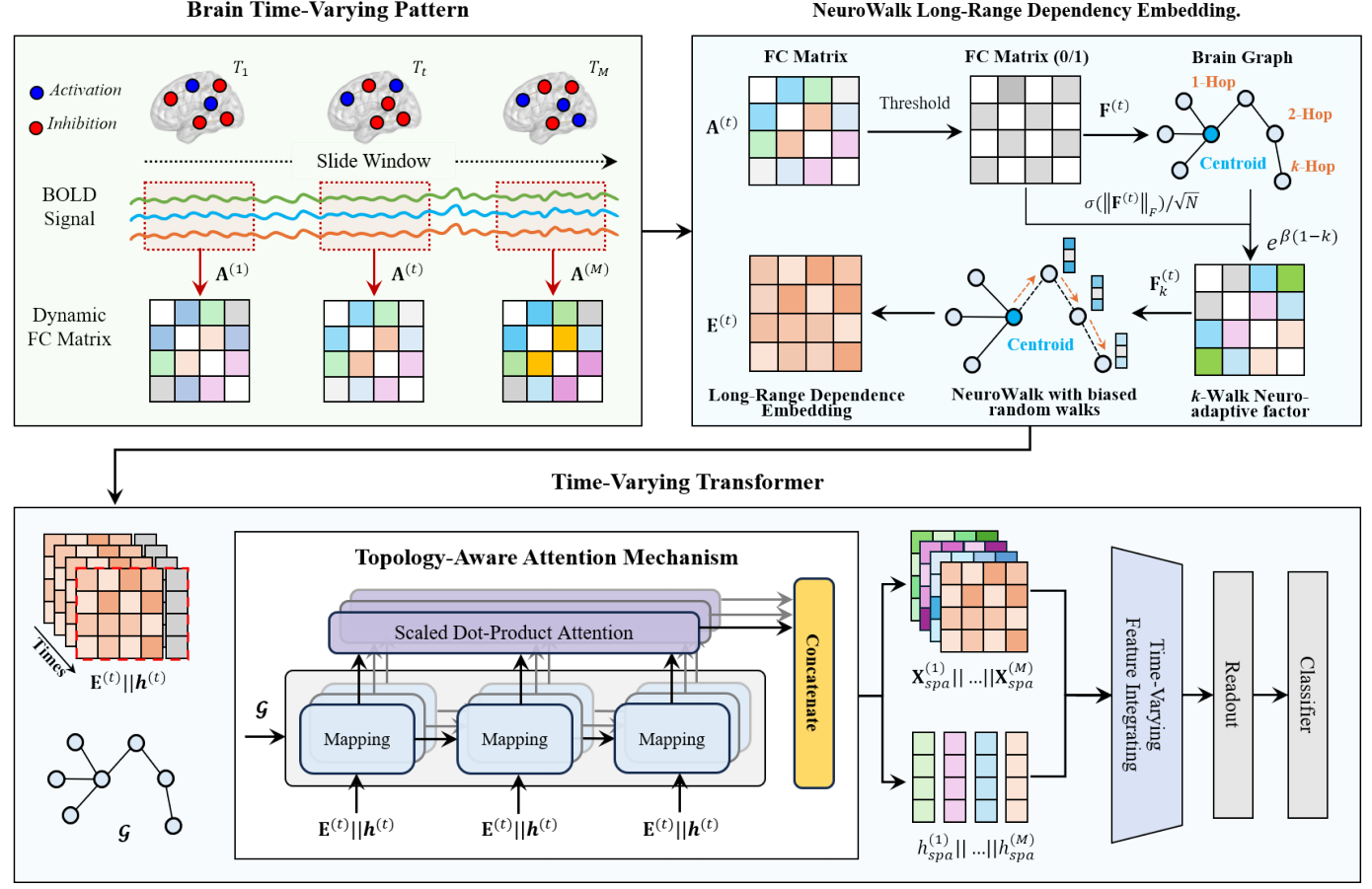 Adaptive Embedding for Long-Range High-Order Dependencies via Time-Varying Transformers on fMRIRundong Xue, Xiangmin Han*, Hao Hu, Zeyu Zhang, Shaoyi Du*, and Yue GaoIn International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025
Adaptive Embedding for Long-Range High-Order Dependencies via Time-Varying Transformers on fMRIRundong Xue, Xiangmin Han*, Hao Hu, Zeyu Zhang, Shaoyi Du*, and Yue GaoIn International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025Dynamic functional brain network analysis using rs-fMRI has emerged as a powerful approach to understanding brain disorders. However, current methods predominantly focus on pairwise brain region interactions, neglecting critical high-order dependencies and time-varying communication mechanisms. To address these limitations, we propose the Long-Range High-Order Dependency Transformer (LHDFormer), a neurophysiologically-inspired framework that integrates multiscale long-range dependencies with time-varying connectivity patterns. Specifically, we present a biased random walk sampling strategy with NeuroWalk kernel-guided transfer probabilities that dynamically simulate multi-step information loss through a k-walk neuroadaptive factor, modeling brain neurobiological principles such as distance-dependent information loss and state-dependent pathway modulation. This enables the adaptive capture of the multi-scale short-range couplings and long-range high-order dependencies corresponding to different steps across evolving connectivity patterns. Complementing this, the time-varying transformer co-embeds local spatial configurations via topology-aware attention and global temporal dynamics through cross-window token guidance, overcoming the single-domain bias of conventional graph/transformer methods. Extensive experiments on ABIDE and ADNI datasets demonstrate that LHDFormer outperforms state-of-the-art methods in brain disease diagnosis. Crucially, the model identifies interpretable high-order connectivity signatures, revealing disrupted long-range integration patterns in patients that align with known neuropathological mechanisms.
@inproceedings{xue2025adaptive, title = {Adaptive Embedding for Long-Range High-Order Dependencies via Time-Varying Transformers on fMRI}, author = {Xue, Rundong and Han, Xiangmin and Hu, Hao and Zhang, Zeyu and Du, Shaoyi and Gao, Yue}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2025}, month = sep, volume = {15971}, pages = {46--55}, doi = {10.1007/978-3-032-05162-2_5}, } - MICCAI 2025
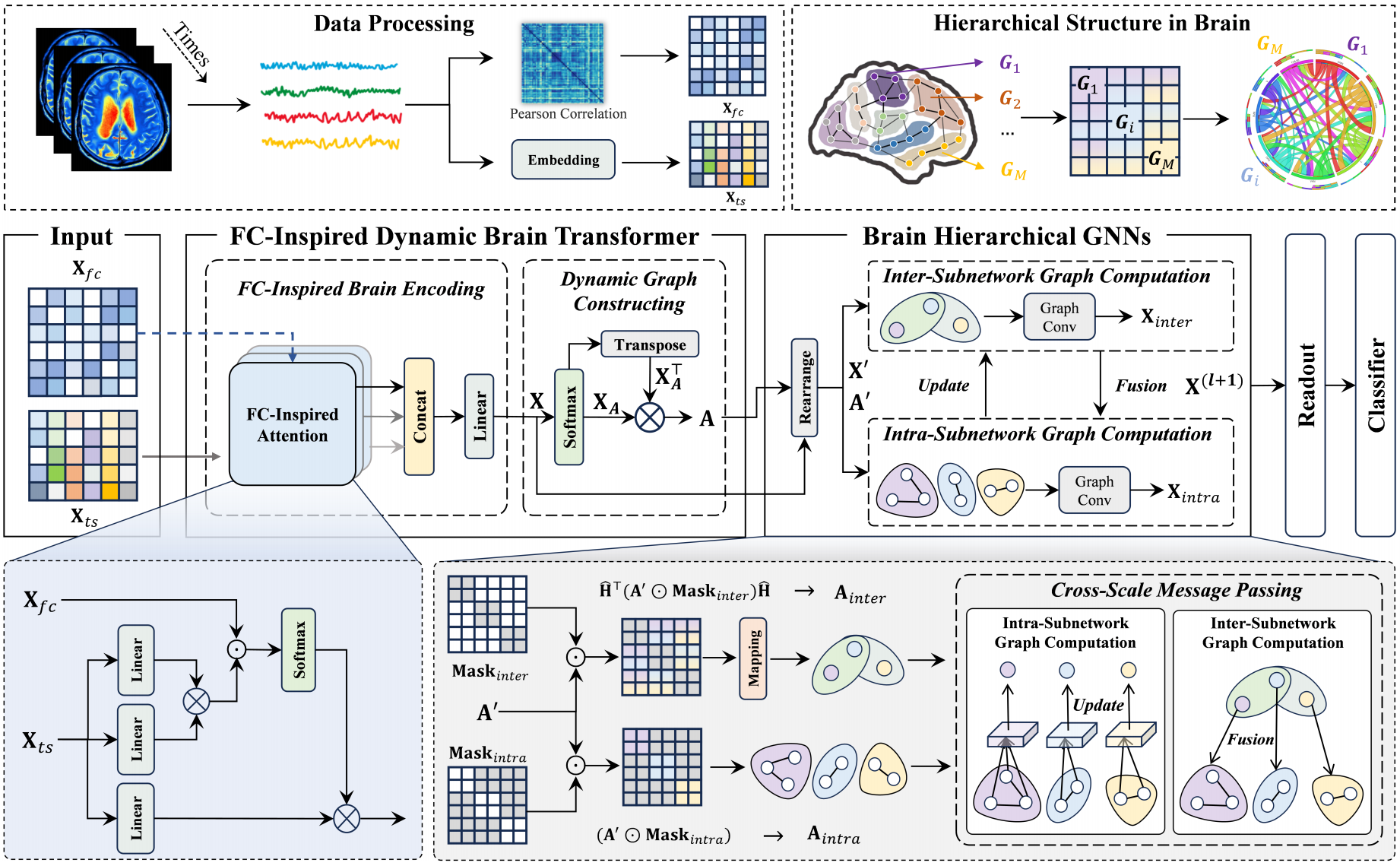 DHGFormer: Dynamic Hierarchical Graph Transformer for Disorder Brain Disease DiagnosisRundong Xue, Hao Hu, Zeyu Zhang, Xiangmin Han*, Juan Wang, Yue Gao, and Shaoyi Du*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025
DHGFormer: Dynamic Hierarchical Graph Transformer for Disorder Brain Disease DiagnosisRundong Xue, Hao Hu, Zeyu Zhang, Xiangmin Han*, Juan Wang, Yue Gao, and Shaoyi Du*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025The functional brain network exhibits a hierarchical characterized organization, balancing localized specialization with global integration through multi-scale hierarchical connectivity. While graph-based methods have advanced brain network analysis, conventional graph neural networks (GNNs) face interpretational limitations when modeling functional connectivity (FC) that encodes excitatory/inhibitory distinctions, often resorting to oversimplified edge weight transformations. Existing methods usually inadequately represent the brain’s hierarchical organization, potentially missing critical information about multi-scale feature interactions. To address these limitations, we propose a novel brain network generation and analysis approach–Dynamic Hierarchical Graph Transformer (DHGFormer). Specifically, our method introduces an FC-driven dynamic attention mechanism that adaptively encodes brain excitatory/inhibitory connectivity patterns into transformer-based representations, enabling dynamic adjustment of the functional brain network. Furthermore, we design hierarchical GNNs that consider prior functional subnetwork knowledge to capture intra-subnetwork homogeneity and inter-subnetwork heterogeneity, thereby enhancing GNN performance in brain disease diagnosis tasks. Extensive experiments on the ABIDE and ADNI datasets demonstrate that DHGFormer consistently outperforms state-of-the-art methods in diagnosing neurological disorders.
@inproceedings{xue2025dhgformer, title = {DHGFormer: Dynamic Hierarchical Graph Transformer for Disorder Brain Disease Diagnosis}, author = {Xue, Rundong and Hu, Hao and Zhang, Zeyu and Han, Xiangmin and Wang, Juan and Gao, Yue and Du, Shaoyi}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2025}, month = sep, volume = {15971}, pages = {268--278}, doi = {10.1007/978-3-032-05162-2_26}, } - IEEE TPAMI, IF 20.8
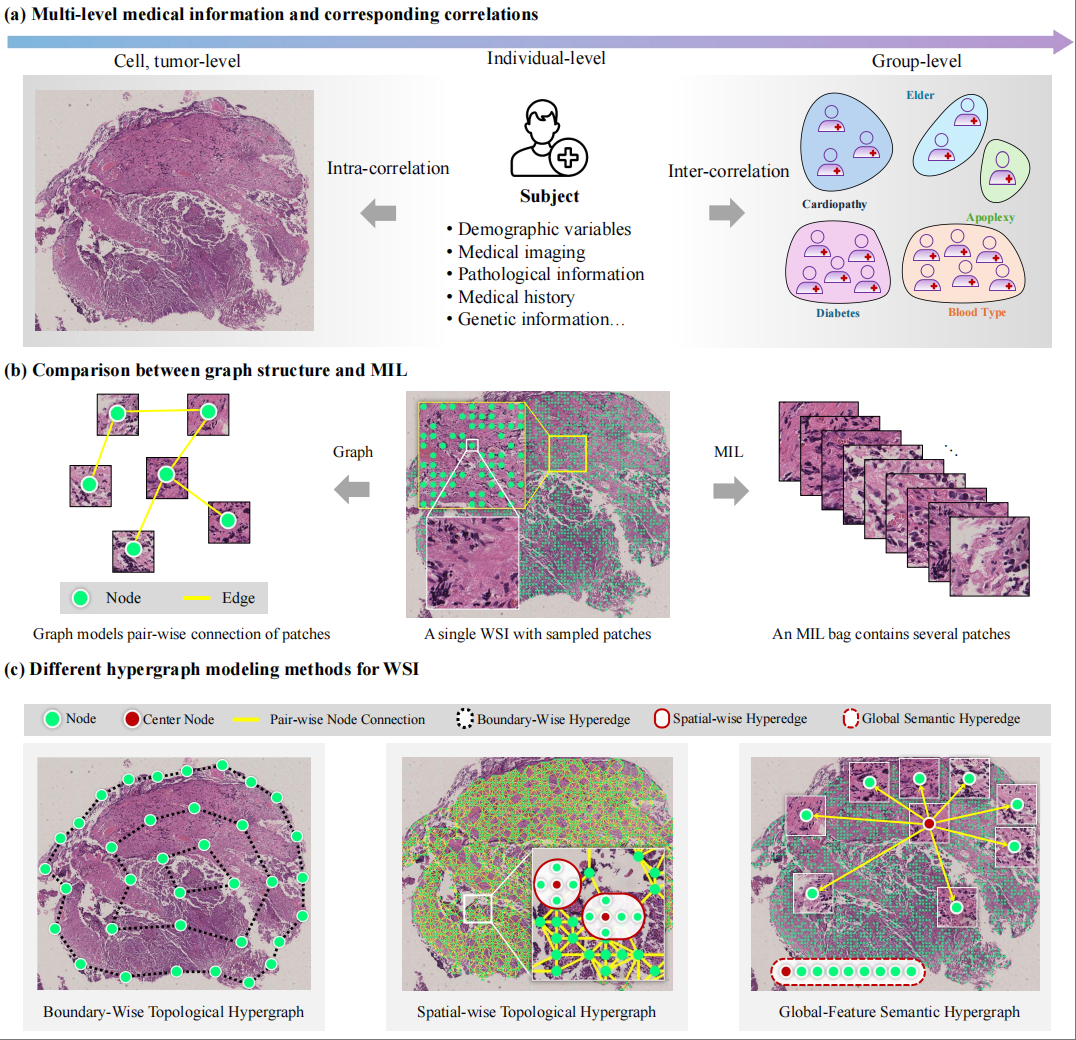 Inter-intra hypergraph computation for survival prediction on whole slide imagesXiangmin Han, Huijian Zhou, Zhiqiang Tian, Shaoyi Du, and Yue Gao*IEEE Transactions on Pattern Analysis and Machine Intelligence, Jul 2025
Inter-intra hypergraph computation for survival prediction on whole slide imagesXiangmin Han, Huijian Zhou, Zhiqiang Tian, Shaoyi Du, and Yue Gao*IEEE Transactions on Pattern Analysis and Machine Intelligence, Jul 2025Survival prediction on histopathology whole slide images (WSIs) involves the analysis of multi-level complex correlations, such as inter-correlations among patients and intra-correlations within gigapixel histopathology images. However, the current graph-based methods for WSI analysis mainly focus on the exploration of pairwise correlations, resulting in the loss of high-order correlations. Hypergraph-based methods can handle such high-order correlations, while existing hypergraph-based methods fail to integrate multi-level high-order correlations into a unified framework, which limits the representation capability of WSIs. In this work, we propose an inter-intra hypergraph computation (I^2HGC) framework to address this issue. The I^2HGC framework implements multi-level hypergraph computation for survival prediction on WSIs, namely intra-hypergraph computation and inter-hypergraph computation. Specifically, the intra-hypergraph computation considers each patch sampled from the histopathology WSI as a vertex of the intra-hypergraph and models the high-order correlations among all patches of an individual WSI in both topology and semantic feature spaces using a hypergraph structure. Then, the intra-hypergraph module generates the intra-embedding and intra-risk for each patient. Subsequently, the inter-hypergraph computation employs these intra-embeddings as features for each patient to form the population-level high-order correlations using data- and knowledge-driven hypergraph modeling strategies. Finally, the intra-risks and the inter-risks are fused for the final survival prediction of each patient. Extensive experimental results on four widely used TCGA carcinoma datasets are presented. We demonstrate that the hypergraph structure captures significantly richer correlations than the graph structure, encompassing all pairwise correlations as well as higher-order interactions through hyperedges. For WSIs with a vast number of pixels and complex correlations, hypergraph-based methods effectively capture topological and semantic information while mitigating the exponential growth of pairwise edges, offering practical advantages for large-scale medical image analysis.
@article{han_2025_inter, title = {Inter-intra hypergraph computation for survival prediction on whole slide images}, author = {Han, Xiangmin and Zhou, Huijian and Tian, Zhiqiang and Du, Shaoyi and Gao, Yue}, journal = {IEEE Transactions on Pattern Analysis and Machine Intelligence}, year = {2025}, month = jul, volume = {47}, number = {7}, pages = {6006--6021}, publisher = {IEEE}, doi = {10.1109/TPAMI.2025.3557391}, } - MedIA, IF: 11.8
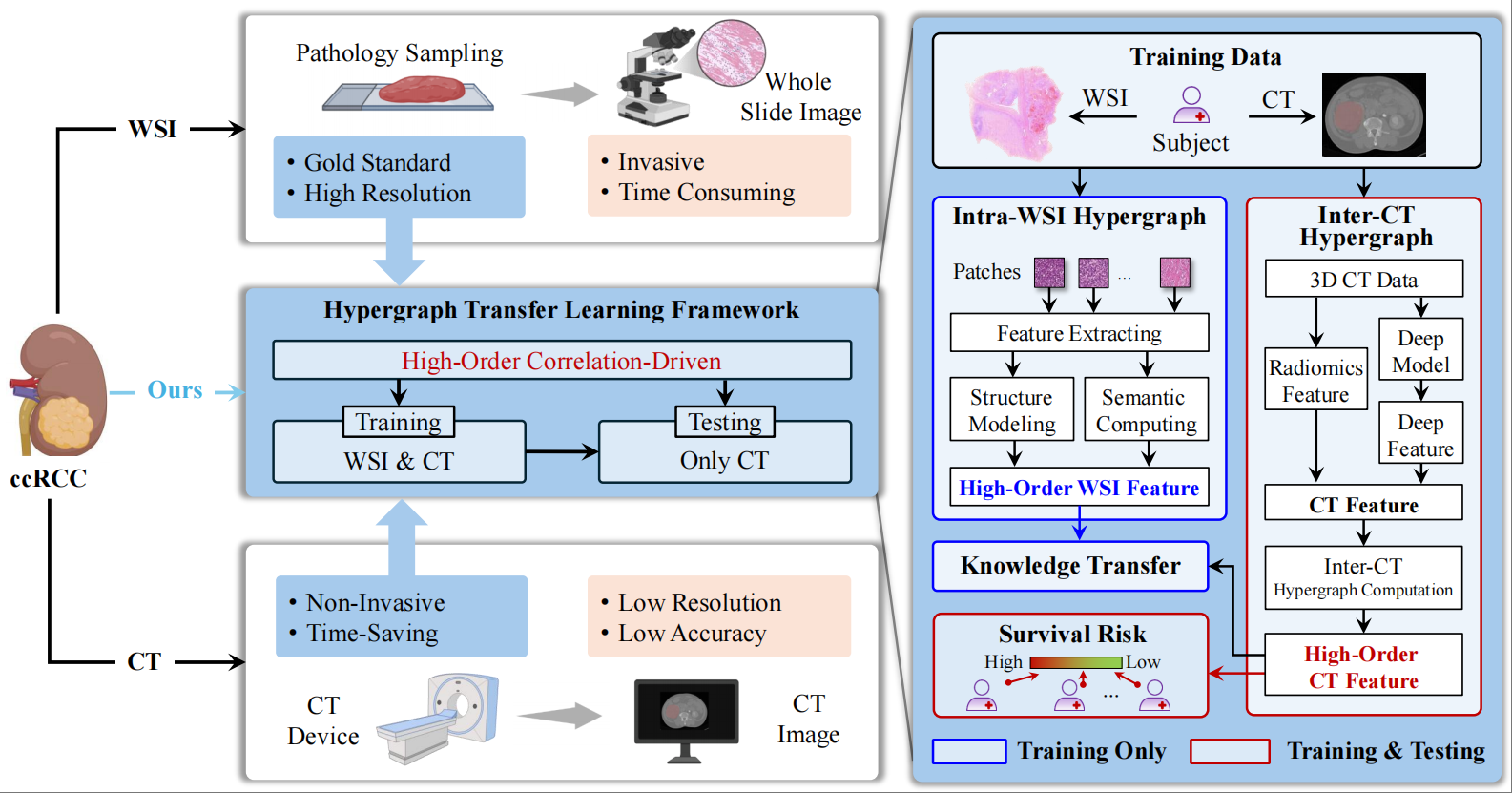 HGTL: A Hypergraph Transfer Learning Framework for Survival Prediction of ccRCCXiangmin Han‡, Wuchao Li‡, Yan Zhang, Pinhao Li, Jianguo Zhu, Tijiang Zhang*, Rongpin Wang*, and Yue Gao*Medical Image Analysis, Oct 2025
HGTL: A Hypergraph Transfer Learning Framework for Survival Prediction of ccRCCXiangmin Han‡, Wuchao Li‡, Yan Zhang, Pinhao Li, Jianguo Zhu, Tijiang Zhang*, Rongpin Wang*, and Yue Gao*Medical Image Analysis, Oct 2025The clinical diagnosis of clear cell renal cell carcinoma (ccRCC) primarily depends on histopathological analysis and computed tomography (CT). Although pathological diagnosis is regarded as the gold standard, invasive procedures such as biopsy carry the risk of tumor dissemination. Conversely, CT scanning offers a non-invasive alternative, but its resolution may be inadequate for detecting microscopic tumor features, which limits the performance of prognostic assessments. To address this issue, we propose a high-order correlation-driven method for predicting the survival of ccRCC using only CT images, achieving performance comparable to that of the pathological gold standard. The proposed method utilizes a cross-modal hypergraph neural network based on hypergraph transfer learning to perform high-order correlation modeling and semantic feature extraction from whole-slide pathological images and CT images. By employing multi-kernel maximum mean discrepancy, we transfer the high-order semantic features learned from pathological images to the CT-based hypergraph neural network channel. During the testing phase, high-precision survival predictions were achieved using only CT images, eliminating the need for pathological images. This approach not only reduces the risks associated with invasive examinations for patients but also significantly enhances clinical diagnostic efficiency. The proposed method was validated using four datasets: three collected from different hospitals and one from the public TCGA dataset. Experimental results indicate that the proposed method achieves higher concordance indices across all datasets compared to other methods.
@article{han_2025_hgtl, title = {HGTL: A Hypergraph Transfer Learning Framework for Survival Prediction of ccRCC}, author = {Han, Xiangmin and Li, Wuchao and Zhang, Yan and Li, Pinhao and Zhu, Jianguo and Zhang, Tijiang and Wang, Rongpin and Gao, Yue}, journal = {Medical Image Analysis}, year = {2025}, volume = {105}, pages = {103700}, month = oct, doi = {10.1016/j.media.2025.103700}, } - IEEE TNNLS, IF 10.2
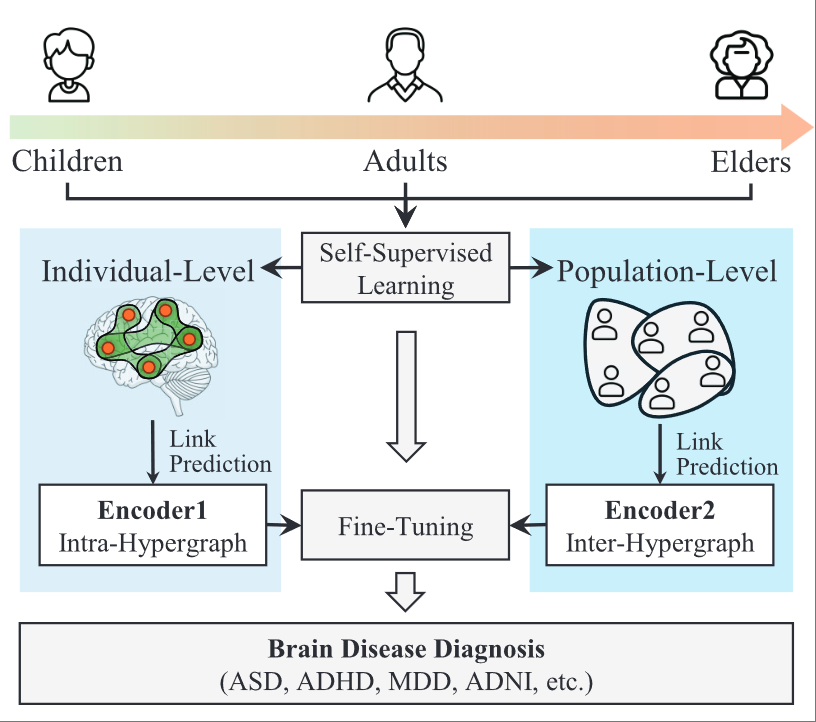 Hypergraph foundation model for brain disease diagnosisXiangmin Han, Rundong Xue, Jingxi Feng, Yifan Feng, Shaoyi Du, Jun Shi*, and Yue Gao*IEEE Transactions on Neural Networks and Learning Systems, Apr 2025
Hypergraph foundation model for brain disease diagnosisXiangmin Han, Rundong Xue, Jingxi Feng, Yifan Feng, Shaoyi Du, Jun Shi*, and Yue Gao*IEEE Transactions on Neural Networks and Learning Systems, Apr 2025The goal of the hypergraph foundation model (HGFM) is to learn an encoder based on the hypergraph computational paradigm through self-supervised pretraining on high-order correlation structures, enabling the encoder to rapidly adapt to various downstream tasks in scenarios, where no labeled data or only a small amount of labeled data are available. The initial exploratory work has been applied to brain disease diagnosis tasks. However, existing methods primarily rely on graph-based approaches to learn low-order correlation patterns between brain regions in brain networks, neglecting the modeling and learning of complex correlations between different brain diseases and patients. This article proposes an HGFM for brain disease diagnosis, which conducts multidimensional pretraining tasks to explore latent cross-dimensional high-order correlation patterns on various brain disease datasets. HGFM is a high-order correlation-driven foundation model for brain disease diagnosis and effectively improves prediction performance. Specifically, HGFM first performs brain functional network link prediction tasks on individual brain networks and group interaction network link prediction tasks on group brain networks, constructing an HGFM for brain disease diagnosis. In downstream tasks, it achieves predictions for different brain disease diagnosis tasks through few-shot learning fine-tuning methods. The proposed method is evaluated on functional magnetic resonance imaging (fMRI) data from 4409 patients across four brain diseases. Results show that it outperforms existing state-of-the-art methods in all brain disease diagnosis tasks, demonstrating its potential value in clinical applications.
@article{han_2025_hypergraph, title = {Hypergraph foundation model for brain disease diagnosis}, author = {Han, Xiangmin and Xue, Rundong and Feng, Jingxi and Feng, Yifan and Du, Shaoyi and Shi, Jun and Gao, Yue}, journal = {IEEE Transactions on Neural Networks and Learning Systems}, year = {2025}, month = apr, volume = {36}, number = {10}, pages = {17702--17716}, publisher = {IEEE}, doi = {10.1109/TNNLS.2025.3554755}, } - ICME 2025
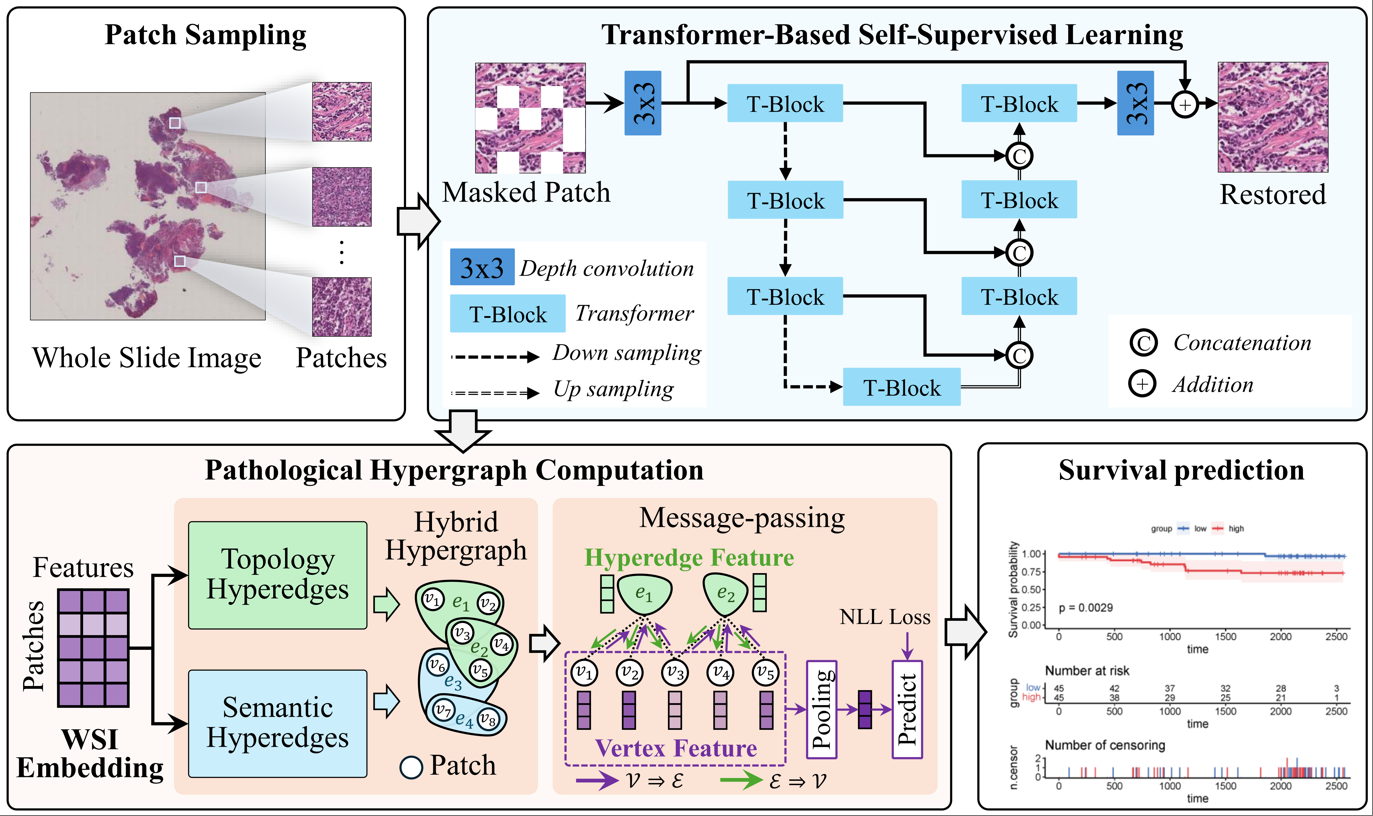 Hypergraph self-supervised learning for survival prediction on whole slide imagesYining Zhao, Hao Liu, Jielong Yan, Yongji Tian, and Xiangmin Han*In IEEE International Conference on Multimedia & Expo, Mar 2025
Hypergraph self-supervised learning for survival prediction on whole slide imagesYining Zhao, Hao Liu, Jielong Yan, Yongji Tian, and Xiangmin Han*In IEEE International Conference on Multimedia & Expo, Mar 2025Existing methods for whole-slide histopathological image analysis commonly depend on patch-level sampling and deep feature extraction based on ImageNet pre-trained models. However, this strategy frequently fails to fully capture the diversity and complex associations within the tissue microenvironment. To address this problem, we propose a HyperGraph Self-supervised Learning (HGSL) method to utilize both local and global information in whole-slide images. Specifically, HGSL consists of two key modules: a Transformer-based self-supervised pretraining (TranSSL) module and a pathology hypergraph neural network (PathHGNN) module. TranSSL uses a mask reconstruction task to mine latent structures and patterns from unlabeled data, thus yielding pathology semantic deep representations. PathHGNN builds a hypergraph structure based on these representations to describe high-order relationships among patches, enabling multi-level feature aggregation and propagation to integrate local microenvironmental information with the overall topological correlations. The proposed HGSL was validated on the TCGA-GBM and TCGA-KIRC datasets, and the experimental results show that HGSL significantly outperforms all existing comparative methods in survival prediction tasks. Furthermore, the KM curve demonstrates that the proposed HGSL significantly distinguishes between high- and low-risk groups, indicating its potential for future clinical applications.
@inproceedings{zhao_hypergraph_2025, title = {Hypergraph self-supervised learning for survival prediction on whole slide images}, author = {Zhao, Yining and Liu, Hao and Yan, Jielong and Tian, Yongji and Han, Xiangmin}, booktitle = {IEEE International Conference on Multimedia & Expo}, year = {2025}, month = mar, } - IEEE TPAMI, IF 18.6
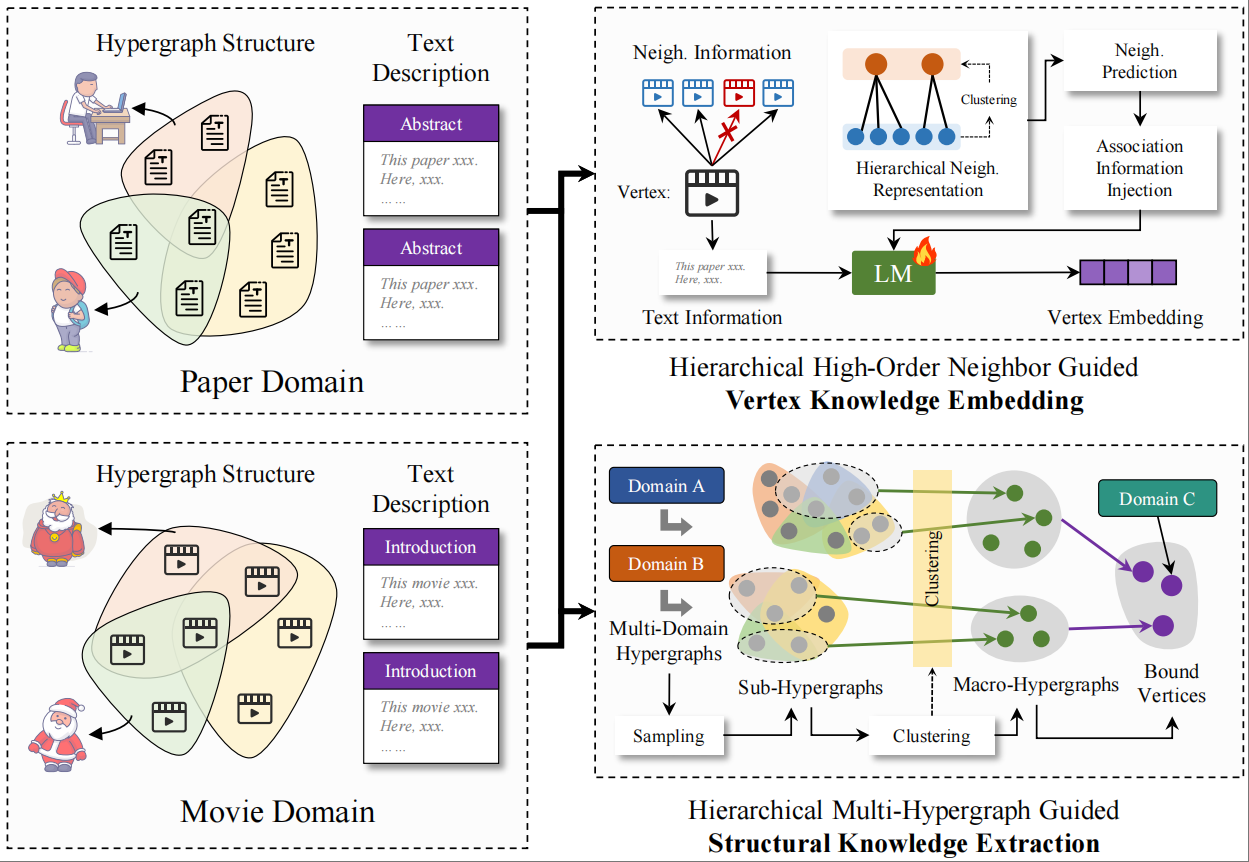 Hypergraph foundation modelYue Gao, Yifan Feng, Shiquan Liu, Xiangmin Han, Shaoyi Du, Zongze Wu*, and Han Hu*IEEE Transactions on Pattern Analysis and Machine Intelligence, Dec 2025
Hypergraph foundation modelYue Gao, Yifan Feng, Shiquan Liu, Xiangmin Han, Shaoyi Du, Zongze Wu*, and Han Hu*IEEE Transactions on Pattern Analysis and Machine Intelligence, Dec 2025 - MICCAI 2025
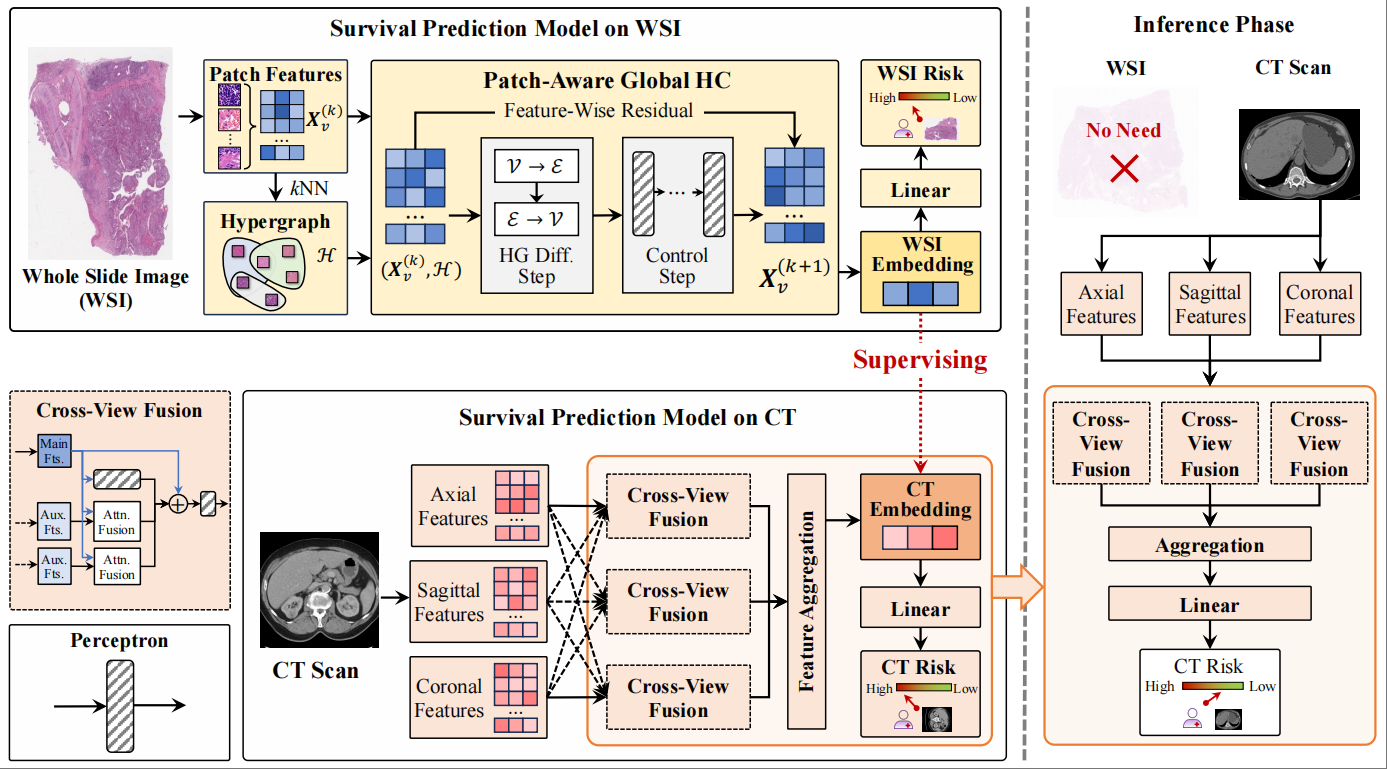 Multimodal Hypergraph Guide Learning for Non-Invasive ccRCC Survival PredictionJielong Yan, Xiangmin Han, Jieyi Zhao, and Yue Gao*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025
Multimodal Hypergraph Guide Learning for Non-Invasive ccRCC Survival PredictionJielong Yan, Xiangmin Han, Jieyi Zhao, and Yue Gao*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025Multimodal medical imaging provides critical data for the early diagnosis and clinical management of clear cell renal cell carcinoma (ccRCC). However, early prediction primarily relies on computed tomography (CT), while whole-slide images (WSI) are often unavailable. Consequently, developing a model that can be trained on multimodal data and make predictions using single-modality data is essential. In this paper, we propose a multimodal hypergraph guide learning framework for non-invasive ccRCC survival prediction. First, we propose a patch-aware global hypergraph computation (PAGHC) module, including a hypergraph diffusion step for capturing correlational structure information and a control step to generate stable WSI semantic embeddings. These WSI semantic embeddings are then used to guide a cross-view fusion method, forming the hypergraph WSI-guided cross-view fusion (HWCVF) to generate CT semantic embeddings, improving single-modality performance in inference. We validate our proposed method on three ccRCC datasets, and quantitative results demonstrate a significant improvement in C-index, outperforming state-of-the-art methods.
@inproceedings{yan2025multimodal, title = {Multimodal Hypergraph Guide Learning for Non-Invasive ccRCC Survival Prediction}, author = {Yan, Jielong and Han, Xiangmin and Zhao, Jieyi and Gao, Yue}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2025}, month = sep, volume = {15971}, pages = {510--520}, doi = {10.1007/978-3-032-05162-2_49}, } - Paper Highlight
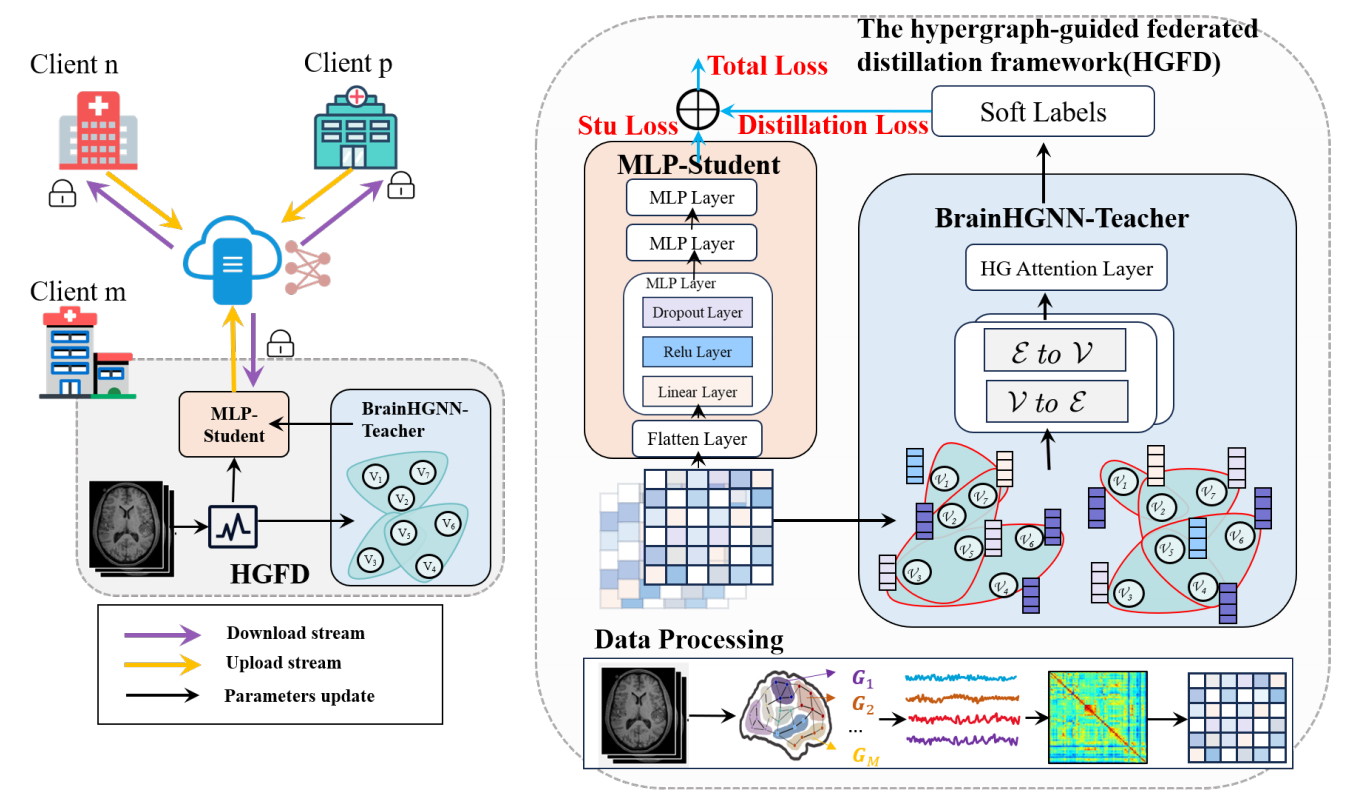 HGFD: Hypergraph Guided Federated Distillation Learning for Efficient and Robust Multi-Center fMRI Data AnalysisTao Jin, Yidan Xu, Yuhan Gao, Xichun Sheng, Chenggang Yan, Yaoqi Sun*, Xiangmin Han, and Yue GaoIn International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025
HGFD: Hypergraph Guided Federated Distillation Learning for Efficient and Robust Multi-Center fMRI Data AnalysisTao Jin, Yidan Xu, Yuhan Gao, Xichun Sheng, Chenggang Yan, Yaoqi Sun*, Xiangmin Han, and Yue GaoIn International Conference on Medical Image Computing and Computer-Assisted Intervention, Sep 2025Multi-center fMRI data analysis faces significant challenges such as data privacy concerns and data integration issues. Federated learning, as an innovative distributed machine learning approach, enables cross-center collaboration by sharing model parameters instead of raw data. However, existing methods often struggle with improving the robustness and inference efficiency of multi-center fMRI data processing. To address these challenges, we propose a novel hypergraph-guided federated distillation framework(HGFD) for multi-center fMRI data analysis. HGFD utilizes a hypergraph structure to model the spatiotemporal features of brain activity, capturing high-order correlations across brain regions. Additionally, a hypergraph knowledge distillation technique is introduced to distill high-order structural information to shallow neural networks, preserving the ability to infer complex relationships while significantly enhancing inference efficiency. In the federated learning process, participating centers only need to share the parameters of their shallow neural networks to a central server. Through parameter aggregation, each center’s shallow network can learn the high-order structural information of other centers. Experiments on multi-center fMRI datasets demonstrate that the proposed method not only improves the robustness and consistency of fMRI-based prediction tasks but also reduces inference time, achieving efficient and accurate predictions while ensuring data privacy.
@inproceedings{jin2025hgfd, title = {HGFD: Hypergraph Guided Federated Distillation Learning for Efficient and Robust Multi-Center fMRI Data Analysis}, author = {Jin, Tao and Xu, Yidan and Gao, Yuhan and Sheng, Xichun and Yan, Chenggang and Sun, Yaoqi and Han, Xiangmin and Gao, Yue}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2025}, month = sep, volume = {15970}, pages = {294--303}, doi = {10.1007/978-3-032-05141-7_29}, } - ICASSP 2025
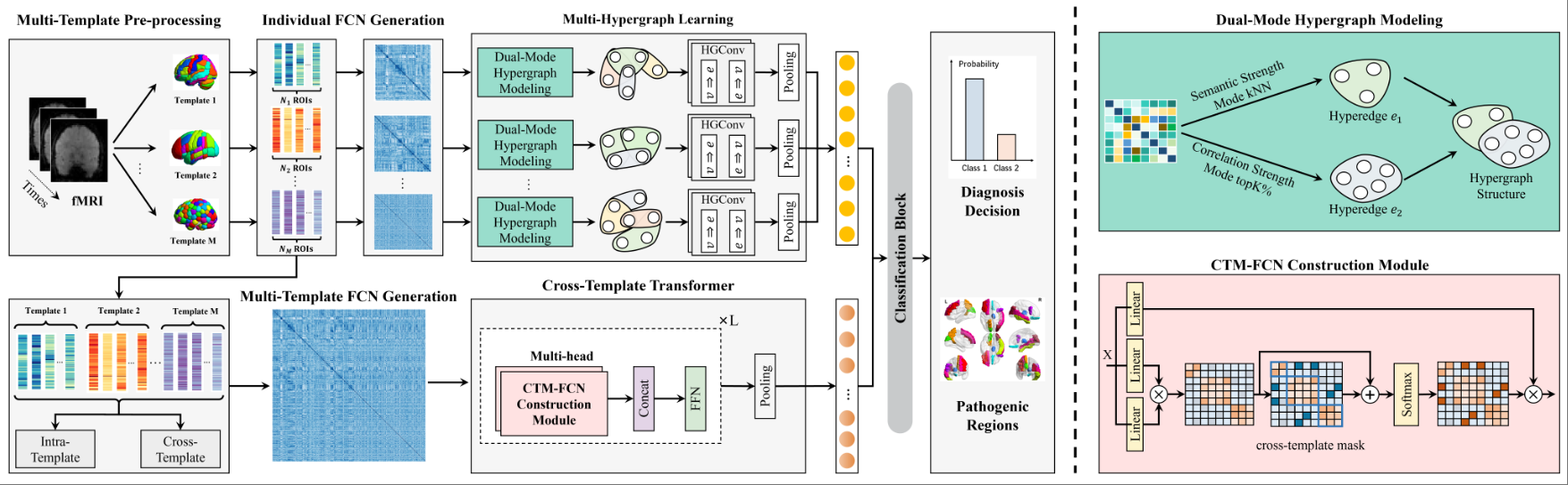 Cross-Template-Based Hypergraph TransformerJingxi Feng, Xiangmin Han, Heming Xu, Juan Wang, Jue Jiang, Shaoyi Du*, and Yue Gao*In IEEE International Conference on Acoustics, Speech and Signal Processing, Sep 2025
Cross-Template-Based Hypergraph TransformerJingxi Feng, Xiangmin Han, Heming Xu, Juan Wang, Jue Jiang, Shaoyi Du*, and Yue Gao*In IEEE International Conference on Acoustics, Speech and Signal Processing, Sep 2025Single-template-based brain functional network analysis methods can provide limited functional connectivity information, which constrains the performance of brain disease diagnosis. Previous works have explored multi-template functional network analysis but failed to integrate the high-order correlation information within templates and the complementary information between templates into a unified relationship strength between nodes, and we extract the high-order correlation information within each template through hypergraph convolution. Secondly, for the analysis of functional connectivity between templates, we propose a cross-template Transformer to capture long-range dependencies between templates. A cross-template mask is applied to focus the model’s attention on important connections between templates, thereby enhancing model robustness. Finally, we progressively fuse the high-order information captured within templates with the global information across templates for downstream classification tasks. The proposed method has been validated on the public ABIDE dataset, and it outperforms existing methods in the ASD diagnosis task.
@inproceedings{feng_2025_cross, title = {Cross-Template-Based Hypergraph Transformer}, author = {Feng, Jingxi and Han, Xiangmin and Xu, Heming and Wang, Juan and Jiang, Jue and Du, Shaoyi and Gao, Yue}, booktitle = {IEEE International Conference on Acoustics, Speech and Signal Processing}, pages = {1--5}, year = {2025}, doi = {10.1109/ICASSP49660.2025.10890417}, }
2024
- MICCAI 2024
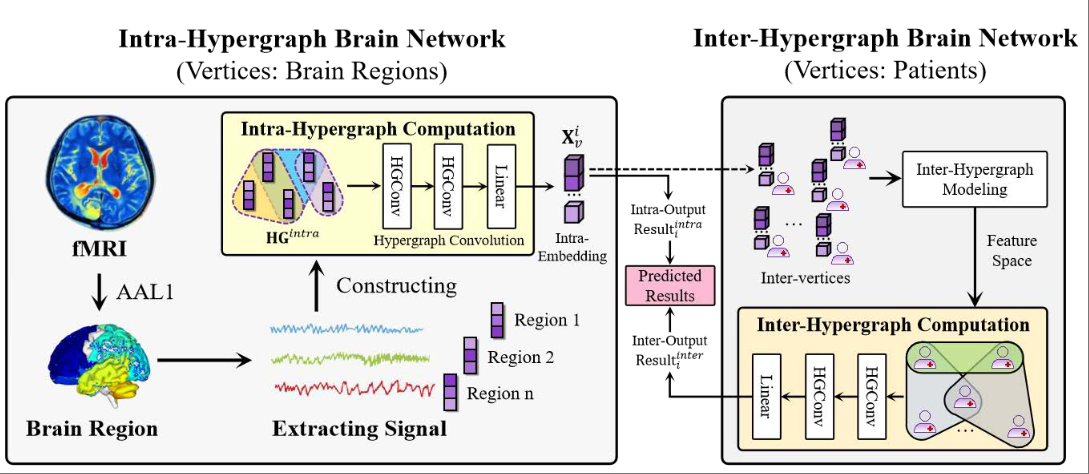 Inter-intra high-order brain network for ASD diagnosis via functional MRIsXiangmin Han, Rundong Xue, Shaoyi Du, and Yue Gao*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Oct 2024
Inter-intra high-order brain network for ASD diagnosis via functional MRIsXiangmin Han, Rundong Xue, Shaoyi Du, and Yue Gao*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Oct 2024Currently in the field of computer-aided diagnosis, graph or hypergraph-based methods are widely used in the diagnosis of neurological diseases. However, existing graph-based work primarily focuses on pairwise correlations, neglecting high-order correlations. Additionally, existing hypergraph methods can only explore the commonality of high-order representations at a single scale, resulting in the lack of a framework that can integrate multi-scale high-order correlations. To address the above issues, we propose an Inter-Intra High-order Brain Network (\mathrmI^2HBN) framework for ASD-assisted diagnosis, which is divided into two parts: intra-hypergraph computation and inter-hypergraph computation. Specifically, the intra-hypergraph computation employs the hypergraph to represent high-order correlations among different brain regions based on fMRI signal, generating intra-embeddings and intra-results. Subsequently, inter-hypergraph computation utilizes these intra-embeddings as features of inter-vertices to model inter-hypergraph that captures the inter-correlations among individuals at the population level. Finally, the intra-results and the inter-results are weighted to perform brain disease diagnosis. We demonstrate the potential of this method on two ABIDE datasets (NYU and UCLA), the results show that the proposed method for ASD diagnosis has superior performance, compared with existing state-of-the-art methods.
@inproceedings{han_inter-intra_2024, title = {Inter-intra high-order brain network for {ASD} diagnosis via functional {MRIs}}, author = {Han, Xiangmin and Xue, Rundong and Du, Shaoyi and Gao, Yue}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2024}, month = oct, volume = {15002}, pages = {216--226}, doi = {10.1007/978-3-031-72069-7_21}, } - MICCAI 2024
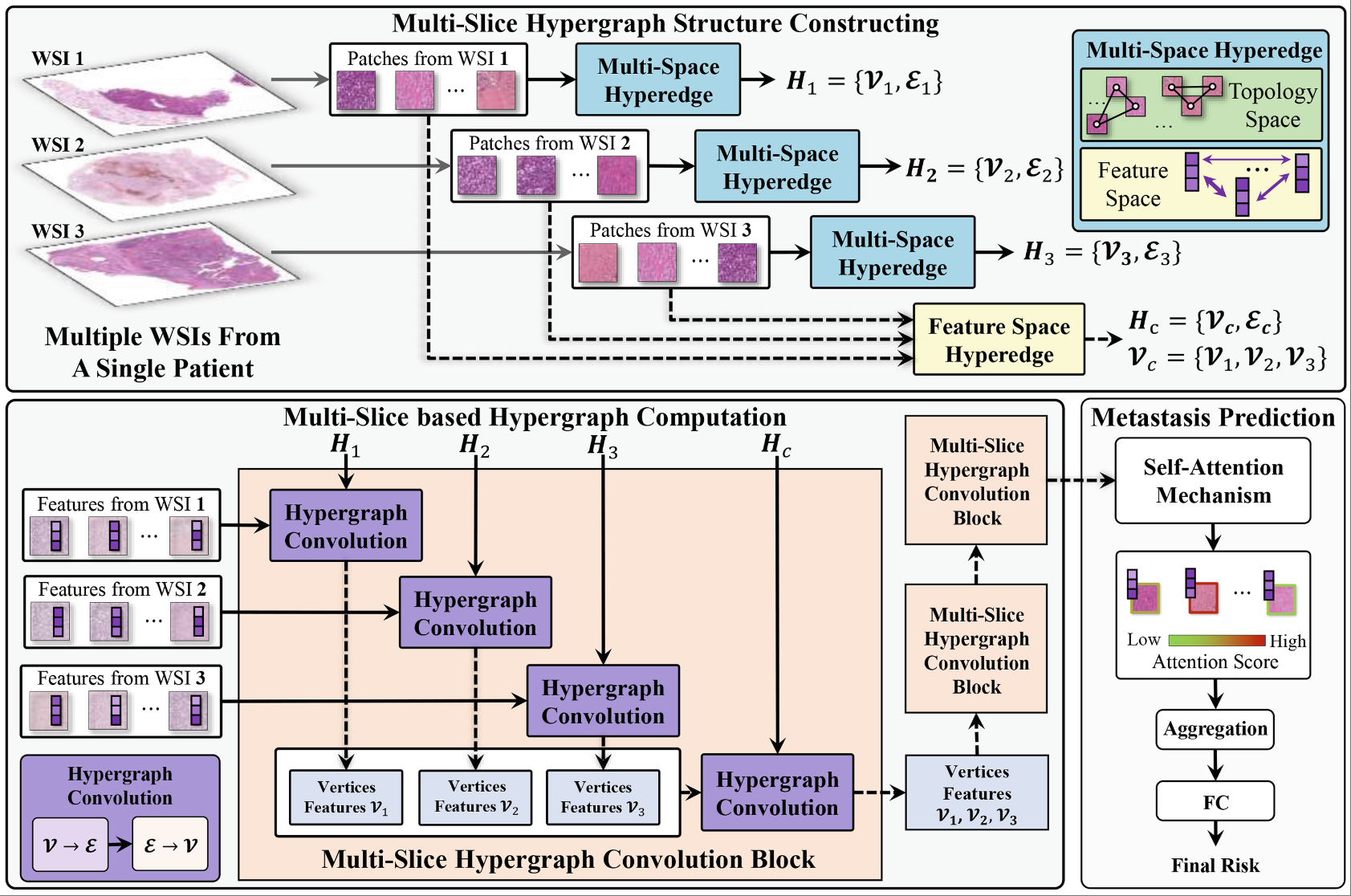 ccRCC metastasis prediction via exploring high-order correlations on multiple WSIsHuijian Zhou, Zhiqiang Tian, Xiangmin Han*, Shaoyi Du, and Yue GaoIn International Conference on Medical Image Computing and Computer-Assisted Intervention, Oct 2024
ccRCC metastasis prediction via exploring high-order correlations on multiple WSIsHuijian Zhou, Zhiqiang Tian, Xiangmin Han*, Shaoyi Du, and Yue GaoIn International Conference on Medical Image Computing and Computer-Assisted Intervention, Oct 2024Metastasis prediction based on gigapixel histopathology whole-slide images (WSIs) is crucial for early diagnosis and clinical decision-making of clear cell renal cell carcinoma (ccRCC). However, most existing methods focus on extracting task-related features from a single WSI, while ignoring the correlations among WSIs, which is important for metastasis prediction when a single patient has multiple pathological slides. In this case, we propose a multi-slice-based hypergraph computation (MSHGC) method for metastasis prediction, which considers the intra-correlations within a single WSI and cross-correlations among multiple WSIs of a single patient simultaneously. Specifically, intra-correlations are captured within both topology and semantic feature spaces, while cross-correlations are modeled between the patches from different WSIs. Finally, the attention mechanism is used to suppress the contribution of task-irrelevant patches and enhance the contribution of task-relevant patches. MSHGC achieves the C-index of 0.8441 and 0.8390 on two carcinoma datasets, outperforming state-of-the-art methods, which demonstrates the effectiveness of the proposed MSHGC.
@inproceedings{zhou_ccrcc_2024, title = {{ccRCC} metastasis prediction via exploring high-order correlations on multiple {WSIs}}, author = {Zhou, Huijian and Tian, Zhiqiang and Han, Xiangmin and Du, Shaoyi and Gao, Yue}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2024}, month = oct, volume = {15005}, pages = {145--154}, doi = {10.1007/978-3-031-72086-4_14}, } - FiM, IF 3.1
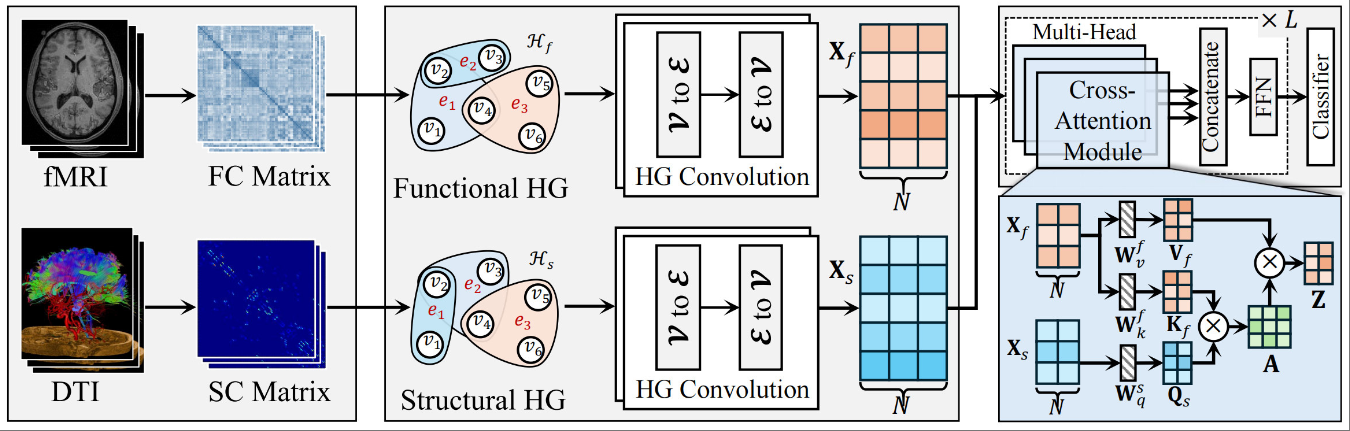 A hypergraph transformer method for brain disease diagnosisXiangmin Han, Jingxi Feng, Heming Xu, Shaoyi Du, and Junchang Li*Frontiers in Medicine, Nov 2024
A hypergraph transformer method for brain disease diagnosisXiangmin Han, Jingxi Feng, Heming Xu, Shaoyi Du, and Junchang Li*Frontiers in Medicine, Nov 2024Objective: To address the high-order correlation modeling and fusion challenges between functional and structural brain networks. Method: This paper proposes a hypergraph transformer method for modeling high-order correlations between functional and structural brain networks. By utilizing hypergraphs, we can effectively capture the high-order correlations within brain networks. The Transformer model provides robust feature extraction and integration capabilities that are capable of handling complex multimodal brain imaging. Results: The proposed method is evaluated on the ABIDE and ADNI datasets. It outperforms all the comparison methods, including traditional and graph-based methods, in diagnosing different types of brain diseases. The experimental results demonstrate its potential and application prospects in clinical practice. Conclusion: The proposed method provides new tools and insights for brain disease diagnosis, improving accuracy and aiding in understanding complex brain network relationships, thus laying a foundation for future brain science research.
@article{han_hypergraph_2024, title = {A hypergraph transformer method for brain disease diagnosis}, author = {Han, Xiangmin and Feng, Jingxi and Xu, Heming and Du, Shaoyi and Li, Junchang}, journal = {Frontiers in Medicine}, year = {2024}, month = nov, volume = {11}, pages = {1496573}, doi = {10.3389/fmed.2024.1496573}, } - Engineering, IF 12.8
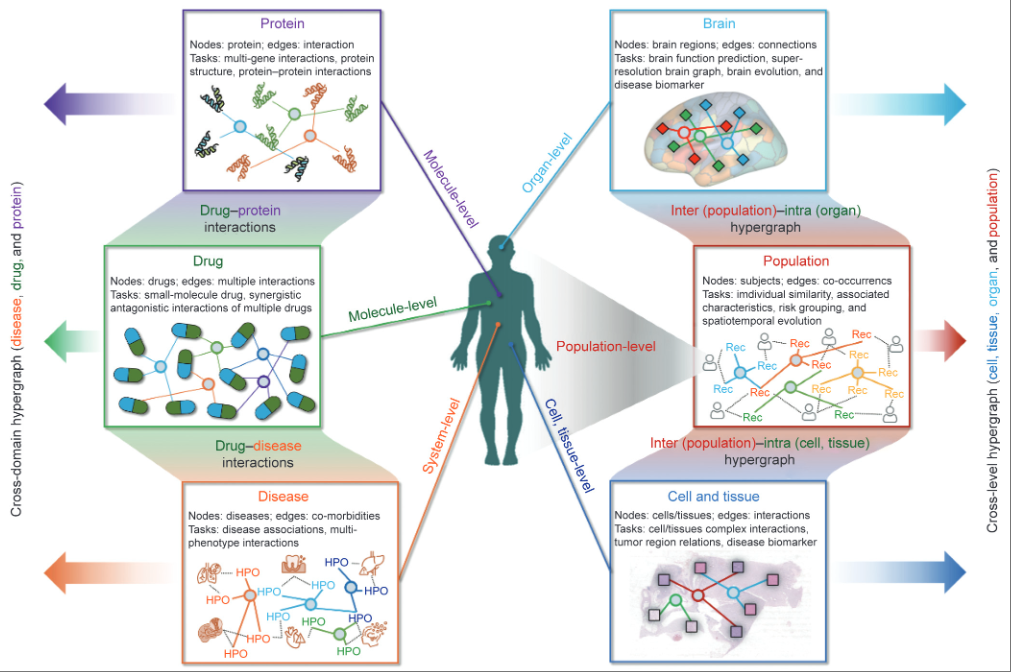 Hypergraph computationYue Gao*, Shuyi Ji, Xiangmin Han, and Qionghai DaiEngineering, Nov 2024
Hypergraph computationYue Gao*, Shuyi Ji, Xiangmin Han, and Qionghai DaiEngineering, Nov 2024Practical real-world scenarios such as the Internet, social networks, and biological networks present the challenges of data scarcity and complex correlations, which limit the applications of artificial intelligence. The graph structure is a typical tool used to formulate such correlations, it is incapable of modeling high-order correlations among different objects in systems; thus, the graph structure cannot fully convey the intricate correlations among objects. Confronted with the aforementioned two challenges, hypergraph computation models high-order correlations among data, knowledge, and rules through hyperedges and leverages these high-order correlations to enhance the data. Additionally, hypergraph computation achieves collaborative computation using data and high-order correlations, thereby offering greater modeling flexibility. In particular, we introduce three types of hypergraph computation methods: ① hypergraph structure modeling, ② hypergraph semantic computing, and ③ efficient hypergraph computing. We then specify how to adopt hypergraph computation in practice by focusing on specific tasks such as three-dimensional (3D) object recognition, revealing that hypergraph computation can reduce the data requirement by 80% while achieving comparable performance or improve the performance by 52% given the same data, compared with a traditional data-based method. A comprehensive overview of the applications of hypergraph computation in diverse domains, such as intelligent medicine and computer vision, is also provided. Finally, we introduce an open-source deep learning library, DeepHypergraph (DHG), which can serve as a tool for the practical usage of hypergraph computation.
@article{gao_2024_hypergraph, title = {Hypergraph computation}, author = {Gao, Yue and Ji, Shuyi and Han, Xiangmin and Dai, Qionghai}, journal = {Engineering}, year = {2024}, volume = {40}, pages = {188--201}, doi = {10.1016/j.eng.2024.04.017}, } - IEEE TMI, IF 8.9
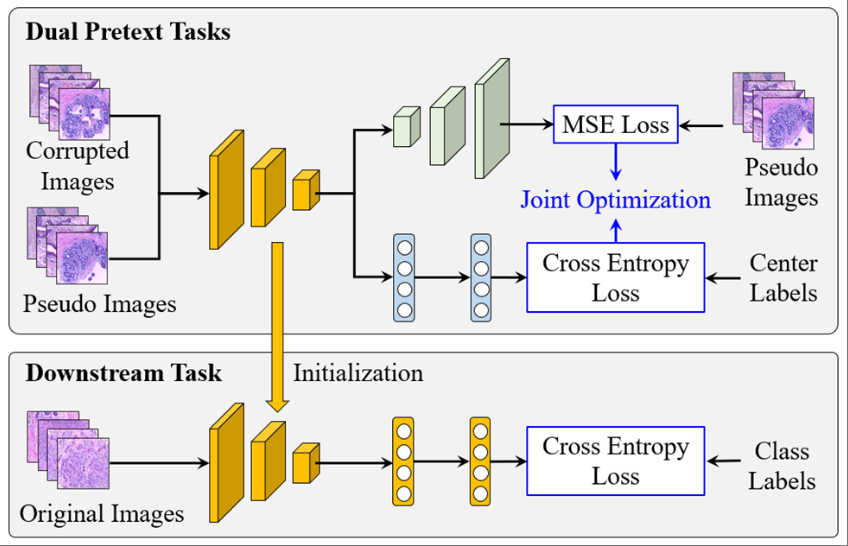 Pseudo-data based self-supervised federated learning for classification of histopathological imagesYuanming Zhang, Zheng Li, Xiangmin Han, Saisai Ding, Juncheng Li, Jun Wang, Shihui Ying, and Jun Shi*IEEE Transactions on Medical Imaging, Mar 2024
Pseudo-data based self-supervised federated learning for classification of histopathological imagesYuanming Zhang, Zheng Li, Xiangmin Han, Saisai Ding, Juncheng Li, Jun Wang, Shihui Ying, and Jun Shi*IEEE Transactions on Medical Imaging, Mar 2024Computer-aided diagnosis (CAD) can help pathologists improve diagnostic accuracy together with consistency and repeatability for cancers. However, the CAD models trained with the histopathological images only from a single center (hospital) generally suffer from the generalization problem due to the straining inconsistencies among different centers. In this work, we propose a pseudo-data based self-supervised federated learning (FL) framework, named SSL-FT-BT, to improve both the diagnostic accuracy and generalization of CAD models. Specifically, the pseudo histopathological images are generated from each center, which contain both inherent and specific properties corresponding to the real images in this center, but do not include the privacy information. These pseudo images are then shared in the central server for self-supervised learning (SSL) to pre-train the backbone of global mode. A multi-task SSL is then designed to effectively learn both the center-specific information and common inherent representation according to the data characteristics. Moreover, a novel Barlow Twins based FL (FL-BT) algorithm is proposed to improve the local training for the CAD models in each center by conducting model contrastive learning, which benefits the optimization of the global model in the FL procedure. The experimental results on four public histopathological image datasets indicate the effectiveness of the proposed SSL-FL-BT on both diagnostic accuracy and generalization.
@article{zhang_pseudo-data_2024, title = {Pseudo-data based self-supervised federated learning for classification of histopathological images}, author = {Zhang, Yuanming and Li, Zheng and Han, Xiangmin and Ding, Saisai and Li, Juncheng and Wang, Jun and Ying, Shihui and Shi, Jun}, journal = {IEEE Transactions on Medical Imaging}, year = {2024}, month = mar, volume = {43}, number = {3}, pages = {902--915}, doi = {10.1109/TMI.2023.3323540}, }
2023
- PR, IF 8.0
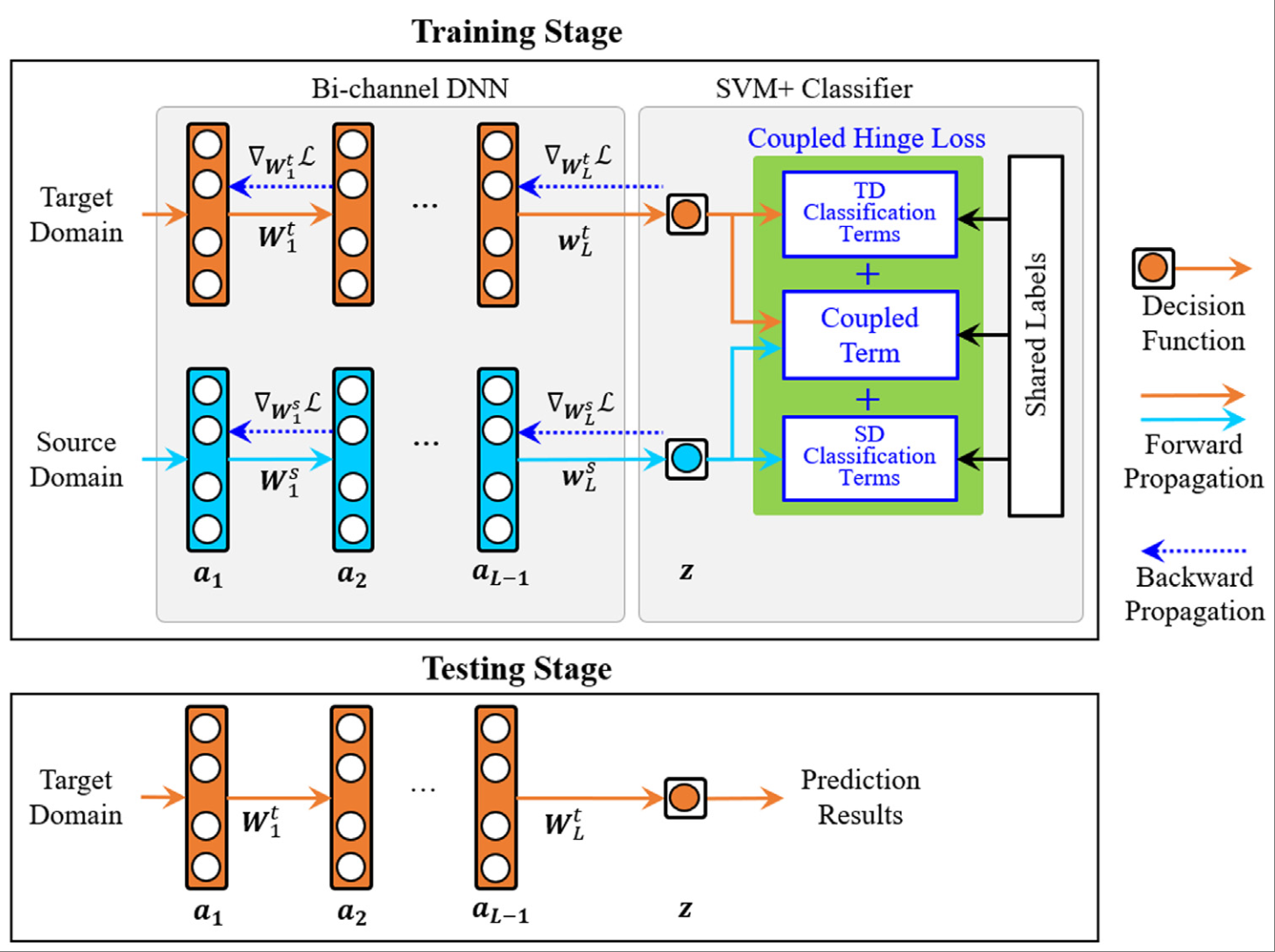 ML-DSVM+: A meta-learning based deep SVM+ for computer-aided diagnosisXiangmin Han, Jun Wang, Shihui Ying, Jun Shi*, and Dinggang Shen*Pattern Recognition, Feb 2023
ML-DSVM+: A meta-learning based deep SVM+ for computer-aided diagnosisXiangmin Han, Jun Wang, Shihui Ying, Jun Shi*, and Dinggang Shen*Pattern Recognition, Feb 2023Transfer learning (TL) can improve the performance of a single-modal medical imaging-based computer-aided diagnosis (CAD) by transferring knowledge from related imaging modalities. Support vector machine plus (SVM+) is a supervised TL classifier specially designed for TL between the paired data in the source and target domains with shared labels. In this work, a novel deep neural network (DNN) based SVM+ (DSVM+) algorithm is proposed for single-modal imaging-based CAD. DSVM+ integrates the bi-channel DNNs and SVM+ classifier into a unified framework to improve the performance of both feature representation and classification. In particular, a new coupled hinge loss function is developed to conduct bidirectional TL between the source and target domains, which further promotes knowledge transfer together with the feature representation under the guidance of shared labels. To alleviate the overfitting caused by the increased parameters in DNNs for limited training samples, the meta-learning based DSVM+ (ML-DSVM+) is further developed, which designs randomly selecting samples from the training data instead of other CAD tasks for meta-tasks. This sampling strategy also can avoid the issue of class imbalance. ML-DSVM+ is evaluated on three medical imaging datasets. It achieves the best results of 88.26±1.40%, 90.45±5.00%, and 87.63±5.56% on accuracy, sensitivity and specificity, respectively, on the Bimodal Breast Ultrasound Image dataset, 90.00±1.05%, 72.55±3.87%, and 96.40±2.26% of the corresponding indices on the Alzheimer’s Disease Neuroimaging Initiative dataset, and 85.76±3.12% of classification accuracy, 88.73±7.22% of sensitivity, and 82.60±1.56% of specificity for the Autism Brain Imaging Data Exchange dataset.
@article{han_ml-dsvm_2023, title = {{ML}-{DSVM}+: {A} meta-learning based deep {SVM}+ for computer-aided diagnosis}, author = {Han, Xiangmin and Wang, Jun and Ying, Shihui and Shi, Jun and Shen, Dinggang}, journal = {Pattern Recognition}, year = {2023}, month = feb, volume = {134}, pages = {109076}, doi = {10.1016/j.patcog.2022.109076}, } - NN, IF 7.8
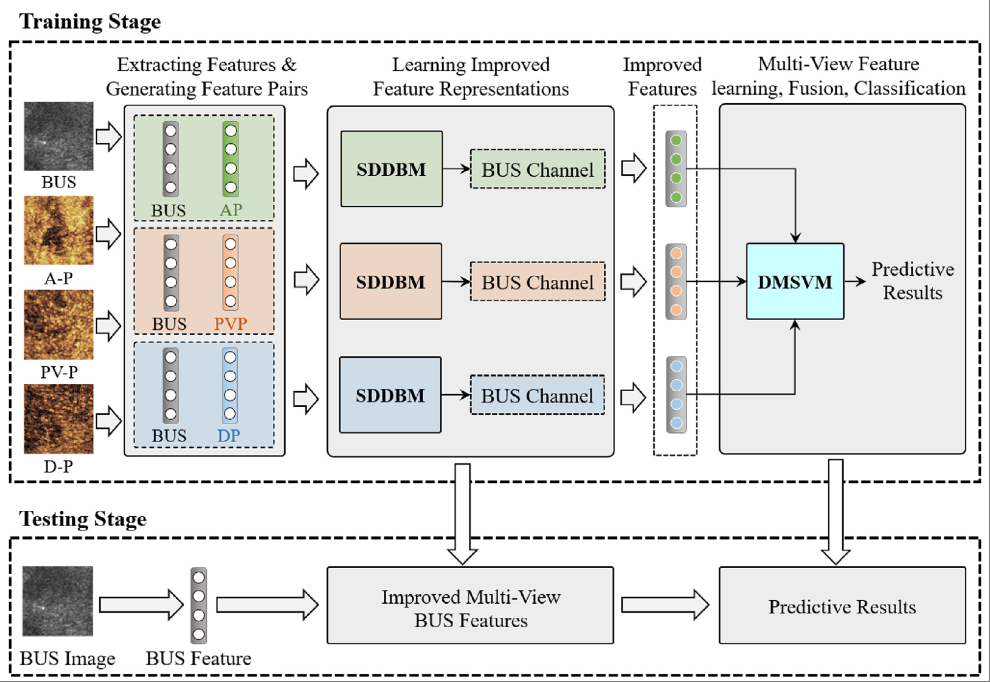 B-mode ultrasound based CAD for liver cancers via multi-view privileged information learningXiangmin Han‡, Bangming Gong‡, Lehang Guo, Jun Wang, Shihui Ying, Shuo Li, and Jun Shi*Neural Networks, Jul 2023
B-mode ultrasound based CAD for liver cancers via multi-view privileged information learningXiangmin Han‡, Bangming Gong‡, Lehang Guo, Jun Wang, Shihui Ying, Shuo Li, and Jun Shi*Neural Networks, Jul 2023B-mode ultrasound-based computer-aided diagnosis model can help sonologists improve the diagnostic performance for liver cancers, but it generally suffers from the bottleneck due to the limited structure and internal echogenicity information in B-mode ultrasound images. Contrast-enhanced ultrasound images provide additional diagnostic information on dynamic blood perfusion of liver lesions for B-mode ultrasound images with improved diagnostic accuracy. Since transfer learning has indicated its effectiveness in promoting the performance of target computer-aided diagnosis model by transferring knowledge from related imaging modalities, a multi-view privileged information learning framework is proposed to improve the diagnostic accuracy of the single-modal B-mode ultrasound-based diagnosis for liver cancers. This framework can make full use of the shared label information between the paired B-mode ultrasound images and contrast-enhanced ultrasound images to guide knowledge transfer It consists of a novel supervised dual-view deep Boltzmann machine and a new deep multi-view SVM algorithm. The former is developed to implement knowledge transfer from the multi-phase contrast-enhanced ultrasound images to the B-mode ultrasound-based diagnosis model via a feature-level learning using privileged information paradigm, which is totally different from the existing learning using privileged information paradigm that performs knowledge transfer in the classifier. The latter further fuses and enhances feature representation learned from three pre-trained supervised dual-view deep Boltzmann machine networks for the classification task. An experiment is conducted on a bimodal ultrasound liver cancer dataset. The experimental results show that the proposed framework outperforms all the compared algorithms with the best classification accuracy of 88.91 ± 1.52%, sensitivity of 88.31 ± 2.02%, and specificity of 89.50 ± 3.12%. It suggests the effectiveness of our proposed MPIL framework for the BUS-based CAD of liver cancers.
@article{han_b-mode_2023, title = {B-mode ultrasound based {CAD} for liver cancers via multi-view privileged information learning}, author = {Han, Xiangmin and Gong, Bangming and Guo, Lehang and Wang, Jun and Ying, Shihui and Li, Shuo and Shi, Jun}, journal = {Neural Networks}, year = {2023}, month = jul, volume = {164}, pages = {369--381}, doi = {10.1016/j.neunet.2023.03.028}, }
2022
- IEEE TMI, IF 10.6
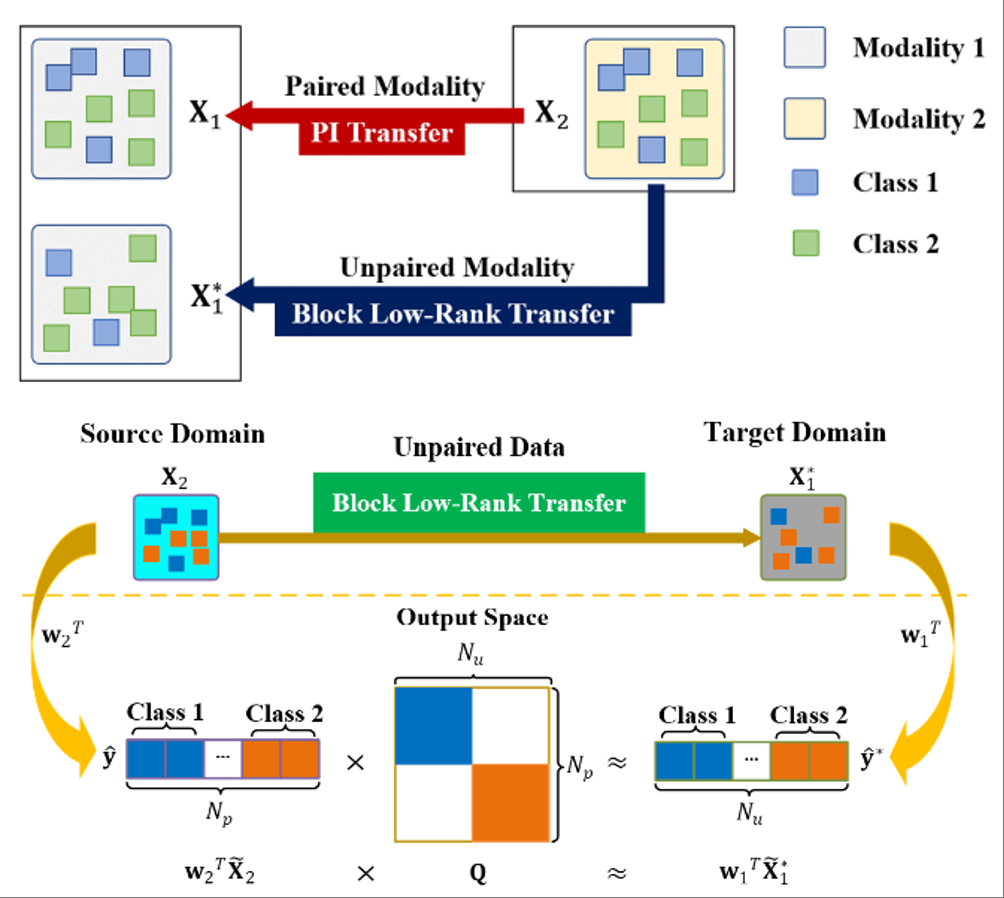 Doubly supervised transfer classifier for computer-aided diagnosis with imbalanced modalitiesXiangmin Han, Xiaoyan Fei, Jun Wang, Tao Zhou, Shihui Ying, Jun Shi*, and Dinggang Shen*IEEE Transactions on Medical Imaging, Aug 2022
Doubly supervised transfer classifier for computer-aided diagnosis with imbalanced modalitiesXiangmin Han, Xiaoyan Fei, Jun Wang, Tao Zhou, Shihui Ying, Jun Shi*, and Dinggang Shen*IEEE Transactions on Medical Imaging, Aug 2022Transfer learning (TL) can effectively improve diagnosis accuracy of single-modal-imaging-based computer-aided diagnosis (CAD) by transferring knowledge from other related imaging modalities, which offers a way to alleviate the small-sample-size problem. However, medical imaging data generally have the following characteristics for the TL-based CAD: 1) The source domain generally has limited data, which increases the difficulty to explore transferable information for the target domain; 2) Samples in both domains often have been labeled for training the CAD model, but the existing TL methods cannot make full use of label information to improve knowledge transfer. In this work, we propose a novel doubly supervised transfer classifier (DSTC) algorithm. In particular, DSTC integrates the support vector machine plus (SVM+) classifier and the low-rank representation (LRR) into a unified framework. The former makes full use of the shared labels to guide the knowledge transfer between the paired data, while the latter adopts the block-diagonal low-rank (BLR) to perform supervised TL between the unpaired data. Furthermore, we introduce the Schatten-p norm for BLR to obtain a tighter approximation to the rank function. The proposed DSTC algorithm is evaluated on the Alzheimer’s disease neuroimaging initiative (ADNI) dataset and the bimodal breast ultrasound image (BBUI) dataset. The experimental results verify the effectiveness of the proposed DSTC algorithm.
@article{han_doubly_2022, title = {Doubly supervised transfer classifier for computer-aided diagnosis with imbalanced modalities}, author = {Han, Xiangmin and Fei, Xiaoyan and Wang, Jun and Zhou, Tao and Ying, Shihui and Shi, Jun and Shen, Dinggang}, journal = {IEEE Transactions on Medical Imaging}, year = {2022}, month = aug, volume = {41}, number = {8}, pages = {2009--2020}, doi = {10.1109/TMI.2022.3152157}, } - IEEE JBHI, IF 7.021
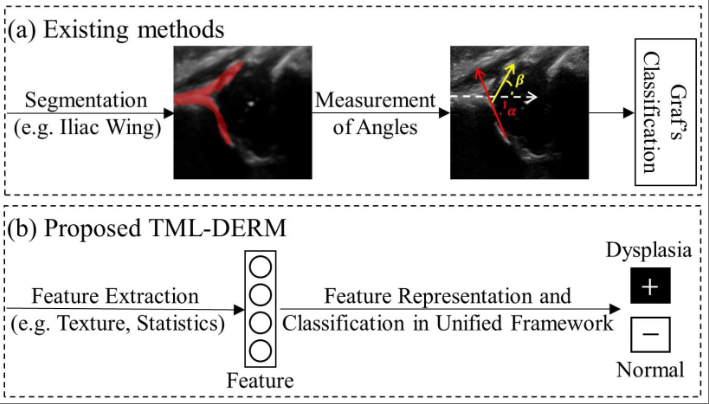 Diagnosis of infantile hip dysplasia with B-mode ultrasound via two-stage meta-learning based deep exclusivity regularized machineBangming Gong, Jing Shi, Xiangmin Han, Huan Zhang, Yuemin Huang, Liwei Hu, Jun Wang, Jun Du, and Jun ShiIEEE Journal of Biomedical and Health Informatics, Jan 2022
Diagnosis of infantile hip dysplasia with B-mode ultrasound via two-stage meta-learning based deep exclusivity regularized machineBangming Gong, Jing Shi, Xiangmin Han, Huan Zhang, Yuemin Huang, Liwei Hu, Jun Wang, Jun Du, and Jun ShiIEEE Journal of Biomedical and Health Informatics, Jan 2022The B-mode ultrasound (BUS) based computer-aided diagnosis (CAD) has shown its effectiveness for developmental dysplasia of the hip (DDH) in infants. In this work, a two-stage meta-learning based deep exclusivity regularized machine (TML-DERM) is proposed for the BUS-based CAD of DDH. TML-DERM integrates deep neural network (DNN) and exclusivity regularized machine into a unified framework to simultaneously improve the feature representation and classification performance. Moreover, the first-stage meta-learning is mainly conducted on the DNN module to alleviate the overfitting issue caused by the significantly increased parameters in DNN, and a random sampling strategy is adopted to self-generate the meta-tasks; while the second-stage meta-learning mainly learns the combination of multiple weak classifiers by a weight vector to improve the classification performance, and also optimizes the unified framework again. The experimental results on a DDH ultrasound dataset show the proposed TML-DERM achieves the superior classification performance with the mean accuracy of 85.89%, sensitivity of 86.54%, and specificity of 85.23%.
@article{gong_diagnosis_2022, title = {Diagnosis of infantile hip dysplasia with {B}-mode ultrasound via two-stage meta-learning based deep exclusivity regularized machine}, author = {Gong, Bangming and Shi, Jing and Han, Xiangmin and Zhang, Huan and Huang, Yuemin and Hu, Liwei and Wang, Jun and Du, Jun and Shi, Jun}, journal = {IEEE Journal of Biomedical and Health Informatics}, year = {2022}, month = jan, volume = {26}, number = {1}, pages = {334--344}, doi = {10.1109/JBHI.2021.3093649}, } - IEEE JBHI, IF 7.021
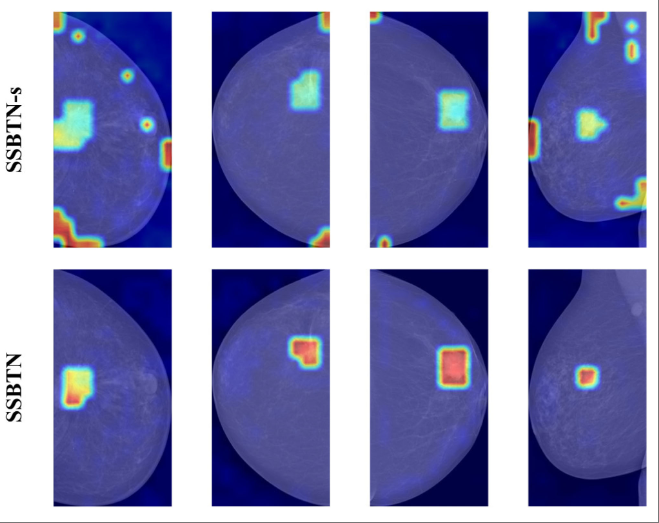 Self-supervised bi-channel transformer networks for computer-aided diagnosisRonglin Gong, Xiangmin Han, Jun Wang, Shihui Ying, and Jun ShiIEEE Journal of Biomedical and Health Informatics, Jul 2022
Self-supervised bi-channel transformer networks for computer-aided diagnosisRonglin Gong, Xiangmin Han, Jun Wang, Shihui Ying, and Jun ShiIEEE Journal of Biomedical and Health Informatics, Jul 2022Self-supervised learning (SSL) can alleviate the issue of small sample size, which has shown its effectiveness for the computer-aided diagnosis (CAD) models. However, since the conventional SSL methods share the identical backbone in both the pretext and downstream tasks, the pretext network generally cannot be well trained in the pre-training stage, if the pretext task is totally different from the downstream one. In this work, we propose a novel task-driven SSL method, namely Self-Supervised Bi-channel Transformer Networks (SSBTN), to improve the diagnostic accuracy of a CAD model by enhancing SSL flexibility. In SSBTN, we innovatively integrate two different networks for the pretext and downstream tasks, respectively, into a unified framework. Consequently, the pretext task can be flexibly designed based on the data characteristics, and the corresponding designed pretext network thus learns more effective feature representation to be transferred to the downstream network. Furthermore, a transformer-based transfer module is developed to efficiently enhance knowledge transfer by conducting feature alignment between two different networks. The proposed SSBTN is evaluated on two publicly available datasets, namely the full-field digital mammography INbreast dataset and the wireless video capsule CrohnIPI dataset. The experimental results indicate that the proposed SSBTN outperforms all the compared algorithms.
@article{gong_self-supervised_2022, title = {Self-supervised bi-channel transformer networks for computer-aided diagnosis}, author = {Gong, Ronglin and Han, Xiangmin and Wang, Jun and Ying, Shihui and Shi, Jun}, journal = {IEEE Journal of Biomedical and Health Informatics}, year = {2022}, month = jul, volume = {26}, number = {7}, pages = {3435--3446}, doi = {10.1109/JBHI.2022.3153902}, }
2021
- PR, IF 7.74
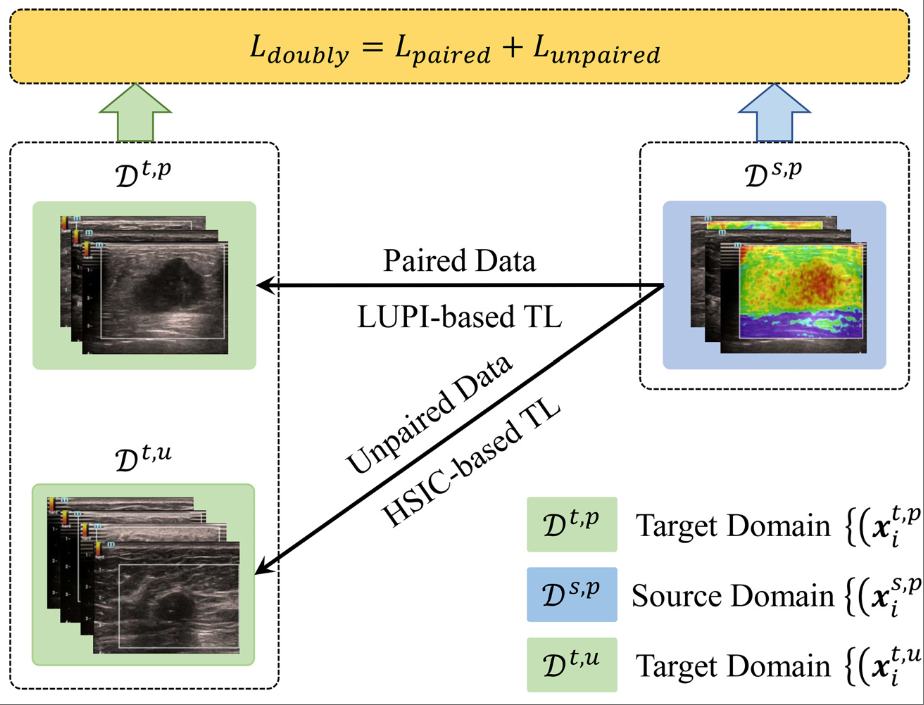 Doubly supervised parameter transfer classifier for diagnosis of breast cancer with imbalanced ultrasound imaging modalitiesXiaoyan Fei, Shichong Zhou, Xiangmin Han, Jun Wang, Shihui Ying, Cai Chang, Weijun Zhou, and Jun ShiPattern Recognition, Dec 2021
Doubly supervised parameter transfer classifier for diagnosis of breast cancer with imbalanced ultrasound imaging modalitiesXiaoyan Fei, Shichong Zhou, Xiangmin Han, Jun Wang, Shihui Ying, Cai Chang, Weijun Zhou, and Jun ShiPattern Recognition, Dec 2021The bimodal ultrasound, namely B-mode ultrasound (BUS) and elastography ultrasound (EUS), provide complementary information to improve the diagnostic accuracy of breast cancers. However, in clinical practice, it is easier to acquire the labeled BUS images than the paired bimodal ultrasound data with shared labels due to the lack of EUS devices, especially in many rural hospitals. Thus, the single-modal BUS-based computer-aided diagnosis (CAD) generally has wide applications. Transfer learning (TL) can promote a BUS-based CAD model by transferring additional knowledge from EUS modality. To make full use of labeled paired bimodal data and the additional single-modal BUS images for knowledge transfer, a novel doubly supervised parameter transfer classifier (DSPTC) is proposed to well handle the TL between imbalanced modalities with the guidance of label information. Specifically, the proposed DSPTC consists of two loss functions corresponding to the paired bimodal ultrasound data with shared labels and the unpaired images with different labels, respectively. The former uses the loss function in the specially designed TL paradigm of support vector machine plus, while the latter adopts the Hilbert-Schmidt Independence Criterion (HSIC) for knowledge transfer between the unpaired images, which consist of the single-modal BUS images and the EUS images from the paired bimodal data. Consequently, the doubly supervised knowledge transfer is implemented by way of parameter transfer in a unified optimization framework. Two experiments are designed to evaluate the proposed DSPTC for the ultrasound-based diagnosis of breast cancers. The experimental results indicate that DSPTC outperforms all the compared algorithms, suggesting its wide potential applications.
@article{fei_doubly_2021, title = {Doubly supervised parameter transfer classifier for diagnosis of breast cancer with imbalanced ultrasound imaging modalities}, author = {Fei, Xiaoyan and Zhou, Shichong and Han, Xiangmin and Wang, Jun and Ying, Shihui and Chang, Cai and Zhou, Weijun and Shi, Jun}, journal = {Pattern Recognition}, year = {2021}, month = dec, volume = {120}, pages = {108139}, doi = {10.1016/j.patcog.2021.108139}, }
2020
- MICCAI 2025
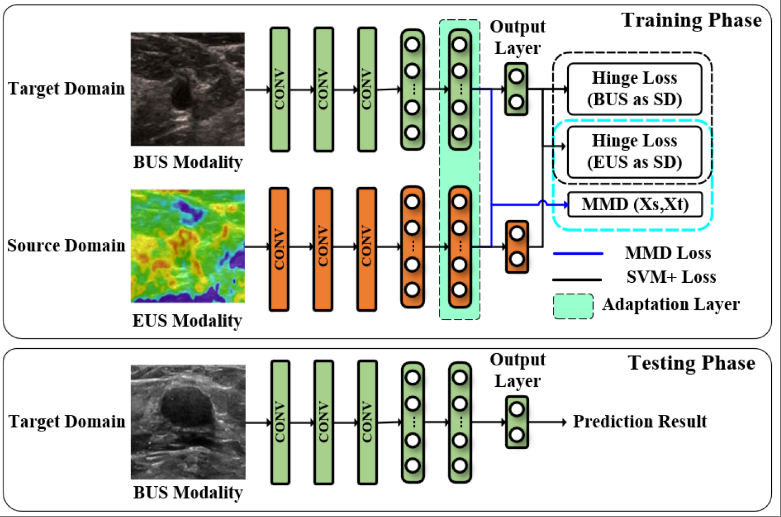 Deep doubly supervised transfer network for diagnosis of breast cancer with imbalanced ultrasound imaging modalitiesXiangmin Han, Jun Wang, Weijun Zhou, Cai Chang, Shihui Ying, and Jun Shi*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Oct 2020
Deep doubly supervised transfer network for diagnosis of breast cancer with imbalanced ultrasound imaging modalitiesXiangmin Han, Jun Wang, Weijun Zhou, Cai Chang, Shihui Ying, and Jun Shi*In International Conference on Medical Image Computing and Computer-Assisted Intervention, Oct 2020Elastography ultrasound (EUS) provides additional bio-mechanical information about lesion for B-mode ultrasound (BUS) in the diagnosis of breast cancers. However, joint utilization of both BUS and EUS is not popular due to the lack of EUS devices in rural hospitals, which arouses a novel modality imbalance problem in computer-aided diagnosis (CAD) for breast cancers. Current transfer learning (TL) pay little attention to this special issue of clinical modality imbalance, that is, the source domain (EUS modality) has fewer labeled samples than those in the target domain (BUS modality). Moreover, these TL methods cannot fully use the label information to explore the intrinsic relation between two modalities and then guide the promoted knowledge transfer. To this end, we propose a novel doubly supervised TL network (DDSTN) that integrates the Learning Using Privileged Information (LUPI) paradigm and the Maximum Mean Discrepancy (MMD) criterion into a unified deep TL framework. The proposed algorithm can not only make full use of the shared labels to effectively guide knowledge transfer by LUPI paradigm, but also perform additional supervised transfer between unpaired data. We further introduce the MMD criterion to enhance the knowledge transfer. The experimental results on the breast ultrasound dataset indicate that the proposed DDSTN outperforms all the compared state-of-the-art algorithms for the BUS-based CAD.
@inproceedings{han_deep_2020, title = {Deep doubly supervised transfer network for diagnosis of breast cancer with imbalanced ultrasound imaging modalities}, author = {Han, Xiangmin and Wang, Jun and Zhou, Weijun and Chang, Cai and Ying, Shihui and Shi, Jun}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, year = {2020}, month = oct, volume = {12266}, pages = {141--149}, doi = {10.1007/978-3-030-59725-2_14}, }